Notebook being hosted here:
http://onsnetwork.org/kubu4/
20081112
Protein Purification (His Column) - Decorin, FST, LAP & Telethonin CONTINUED (from yesterday)
Columns were prepared according to Invitrogen ProBond protocol following the Native Binding/Elution conditions. Samples were loaded onto the prepared columns and incubated at RT for 60mins. on the rocker. The supernatant following this step was saved and stored at 4C. The columns were washed according to the protocol and the washes were collected and combined into a 50mL conical and stored at 4C. The samples were eluted according to the protocl and 8 x 1mL fractions were collected for each protein and stored @ 4C.
Co-IP - MSTN1b + O.mykiss red muscle extract CONTINUED (from yesterday)
The bands were treated according to the Goodlett Lab protocol and stored @ -80C.PCR - Y2H colony screen: Trout muscle cell culture library + MSTN1b mating CONTINUED (from yesterday)
Gel #1. Original image here.
Gel #2. Original image here.
Results: Bands were excised and purified with Millipore spin columns.
20081111
PCR - Y2H colony screen: Trout muscle cell culture library + MSTN1b mating
Colonies from plates P3 and P4 were PCR'd. PCR was run O/N. Plate layout and PCR set up here.Protein Purification (His Column) - Decorin, FST, LAP & Telethonin (from 20081031)
These four preps were sent through a YM-3 Centricon filter by spinning 4000RPM @ 4C for 30mins. Retentate was mixed with an appropriate volume of 5X Native Binding Buffer w/o Imidazole (Invitrogen ProBond protocol) to result in a 1X solution. Volume was adjusted to 8mL with 1X Native Binding Buffer w/o Imidazole. Samples were stored @ 4C O/N.Co-IP - MSTN1b + O.mykiss red muscle extract CONTINUED (from 20081106)
Destained bands were treated according to Goodlett Lab Protein Digestion protocol and were incubated O/N with trypsin.20081107
PCR - Y2H colony screen: Trout muscle cell culture library + MSTN1b mating CONTINUED (from yesterday)
Purified PCR products were sent for sequencing. See sequence log for plate layout/primers.
20081106
Co-IP - MSTN1b + O.mykiss red muscle extract CONTINUED (from 20081104)
Samples were treated and eluted according to Pierce anti-myc co-IP kit protcol. Initial column flow-through from each prep was saved. 1uL of the flow-through samples was added to 25uL of 2x Pierce sample buffer and heated @ 95C for 10mins along with the columns during the elution step. 1uL of 2-ME was added to every sample and the samples were loaded onto a Pierce 4-20% tris-hepes SDS/PAGE gel. 10uL of SeeBlue ladder was loaded. Gel was run @ 150V for 40mins. Gel was silver stained according to Invitrogen SilverQuest kit protocol.Original picture here.
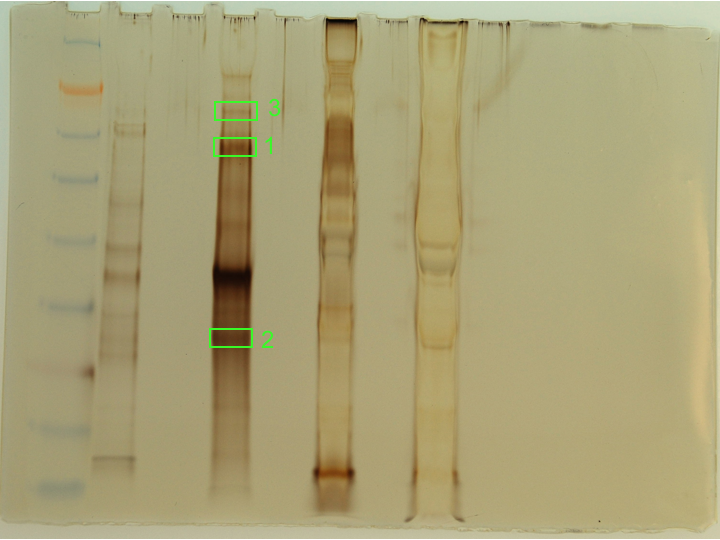
Lane 1 = SeeBlue ladder
Lane 2 = Empty
Lane 3 = <50kDa Flow-through
Lane 4 = Empty
Lane 5 = <50kDa elution
Lane 6 = Empty
Lane 7 = >50kDa Flow-through
Lane 8 = Empty
Lane 9 = >50kDa elution
Results: Bands boxed in green were excised and destained according to Invitrogen SilverQuest kit protocol and stored @ -20C until ready for digestion and submission for sequencing. The bands were chosen based on their intensity and, thus, their likelihood of having sufficient quantities of protein for mass spec analysis. The >50kDa elution sample contained a great deal of insoluble debris and that may help explain why that sample looks so strange (Lane 9).
PCR - Y2H colony screen: Trout muscle cell culture library + MSTN1b mating CONTINUED (from yesterday)
Remaining samples (P3, colonies #6-8 and neg. control) were run on a gel.
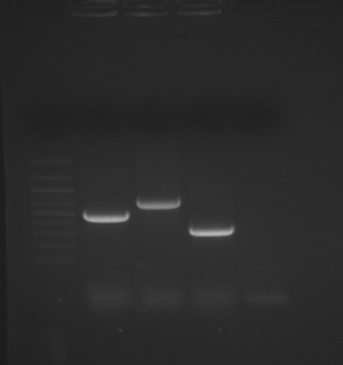
Bands were excised and purified with Millipore spin columns. See yesterday for labeling scheme.
20081105
PCR - Y2H colony screen: Trout muscle cell culture library + MSTN1b mating
Colonies from plates P1, P2 and P2 were PCR'ed and run on gels. PCR set up here. Plate layout here.Gel #1. Original picture here.
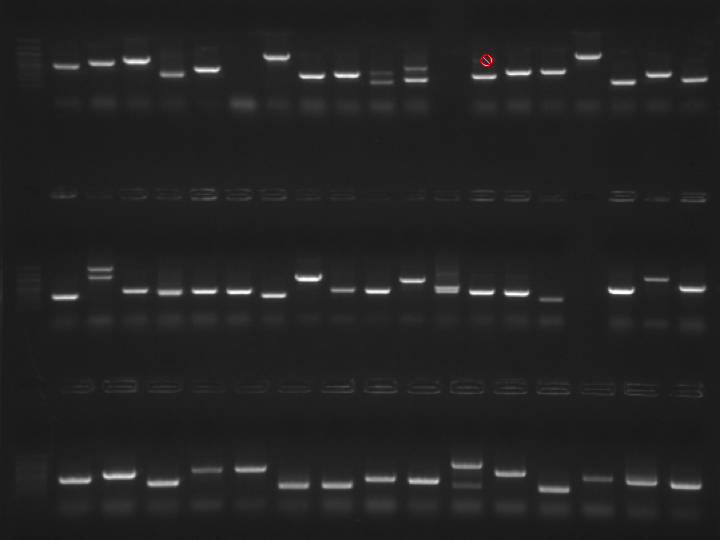
Top Row - 100bp Ladder, P1 Colonies #1-19
Middle Row - 100bp Ladder, P1 Colonies #20-34, 36-38 (#35 not loaded due to evaporated sample)
Bottom Row - 100bp Ladder, Colony #39-45, P2 Colonies 1-8
Gel #2. Original picture here.
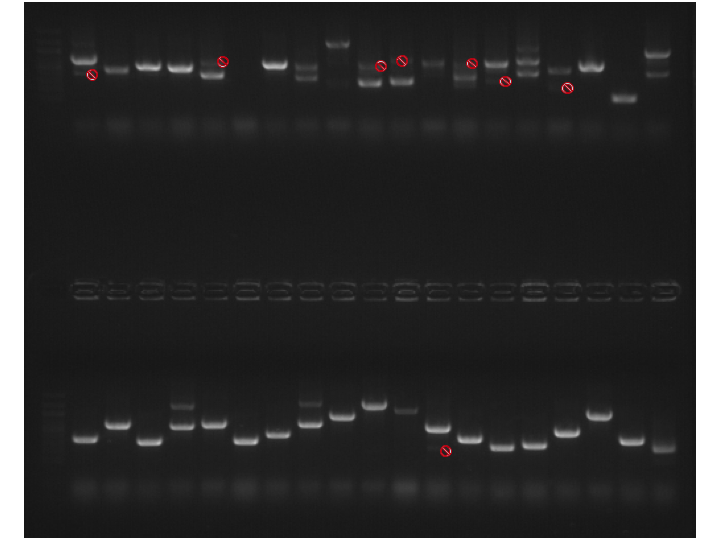
Top Row - 100bp Ladder, P2 Colonies #9-27
Bottom Row - 100bp Ladder, P2 Colonies #28-42, P3 Colonies #2-5
Results: PCRs look good. All bands that are NOT marked with a red "Do Not Enter" symbol were not excised. All others were excised and purified using Millipore spin columns. Cut out bands were labeled with the plate # (P1, P2 or P3), the colony # and then the band, where multiple bands are labeled with 1 for the largest band, 2 for the next largest, and so on. Example for Gel 2, Lane 9: P2.16-1 and P2.16-2.
PCR - Y2H colonies: Spot check of red muscle library + MSTN1b cotrans
A selection of colonies (Plate Q29 #s 1-18) that were sent for sequencing (see 20081031) will be PCR'ed under the same conditions as those for the sequencing run and run on a gel to assess results. PCR set up here.Original picture here.

Results: Nearly every lane has multiple bands which could explain why the sequencing results were crappy. Bands surrounded by green boxes were excised and purified using Millipore spin columns. Cut out bands were labeled with the plate # (Q29), the colony # and then the band, where multiple bands are labeled with 1 for the largest band, 2 for the next largest, and so on. Example for lane #2: Q29.1-1 and Q29.1-2.
20081104
Co-IPs - MSTN1b active + possible targets CONTINUED (from yesterday)
Samples were treated and eluted according to Pierce anti-myc co-IP kit protcol. Initial column flow-through from each prep was saved. 1uL of the flow-through samples was added to 25uL of 2x Pierce sample buffer and heated @ 95C for 10mins along with the columns during the elution step. 1uL of 2-ME was added to every sample and the samples were loaded onto a Pierce 4-20% tris-hepes SDS/PAGE gel. 10uL of SeeBlue ladder was loaded as well. Gel was run @ 150V for 40mins. Gel was silver stained according to Invitrogen SilverQuest kit protocol.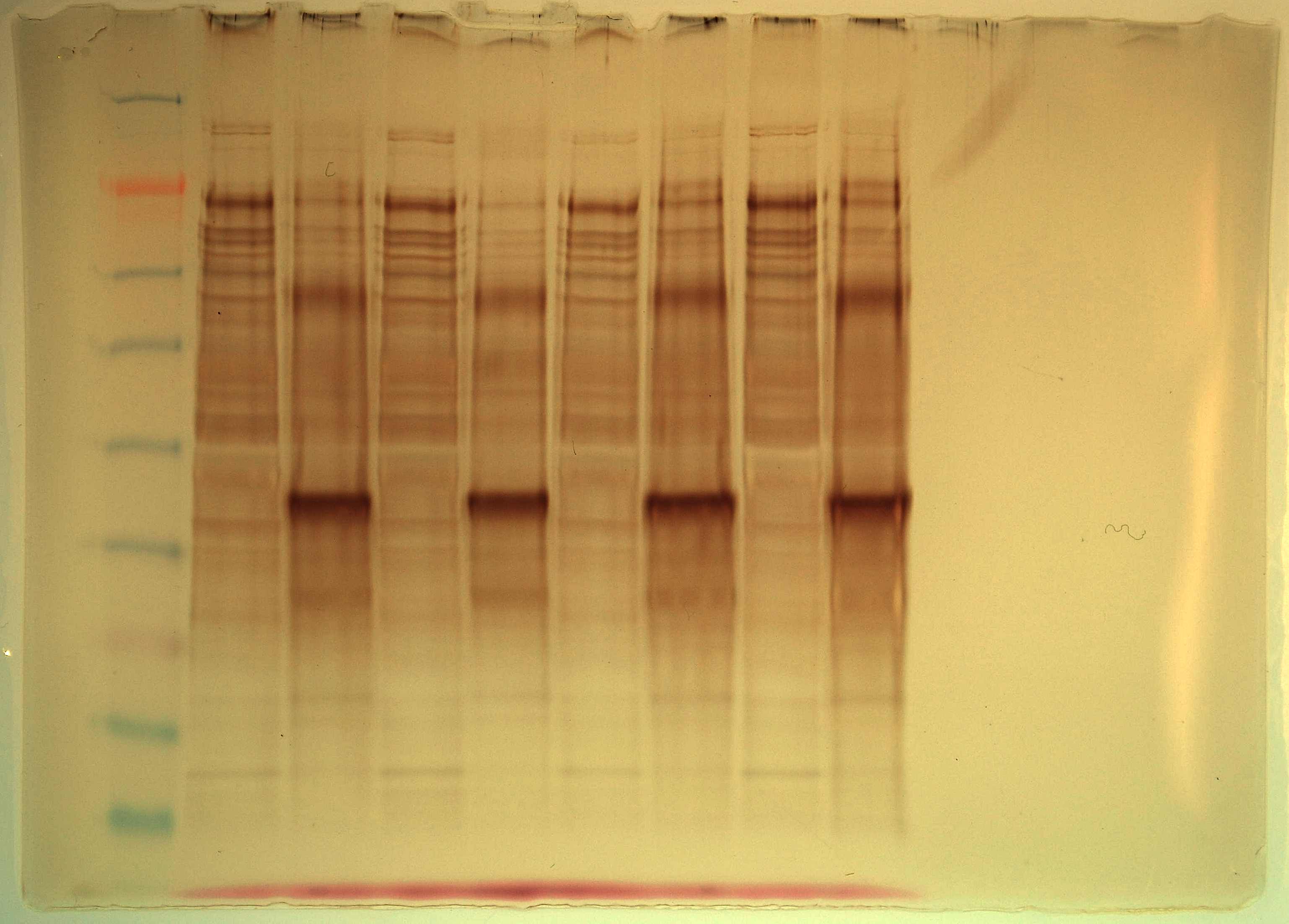
Lane 1 = SeeBlue ladder
Lane 2 = Decorin flow-through
Lane 3 = Decorin elution
Lane 4 = FST flow-through
Lane 5 = FST elution
Lane 6 = LAP flow-through
Lane 7 = LAP elution
Lane 8 = Telethonin flow-through
Lane 9 = Telethonin elution
Results: The dark, prominent bands in each of the elutions are MSTN1b. The higher molecular weight band is likely a MSTN1b dimer. Otherwise, it appears as though there is no detectable protein that co-IP with MSTN1b. Will try to purify these proteins using a Nickel column to bind to the His tag that should be on these recombinant proteins. Then will attempt co-IP on purified protein samples.
20081103
2YH: Trout muscle cell library mating CONTINUED (from 20081031)
Yeast colonies were restreaked onto QDO plates (100mm) by Steven and Mac. Plates were labeled with Pn and incubated @ 30C.
Co-IPs - MSTN1b active + possible targets
700uL of dialyzed (against 1x TBS) Decorin, FST, LAP and Telethonin bacterial lysates from 20081031 were combined in separate co-IP columns with 100uL of MSTN1b active and 10uL of anti-myc agarose slurry according to the Pierce anti-myc coIP Kit protocol.. These were incubated O/N @ 4C w/end-over-end rocking.20081031
pBAD Expression - Dialysis of Decorin, FST, LAP & Telethonin CONTINUED (from yesterday)
1X TBS was changed twice during the day and incubated at 4C with stirring over the weekend. Co-IPs with MSTN1b active form will be performed on Monday. Samples will be transferred to 15mL conical tubes and stored @ 4C. Insoluble and soluble portions of these preps will be combined into a single tube.Y2H: Trout muscle cell library mating CONTINUED (from yesterday)
Cells were prepared according to Clontech Matchmaker protocol for library mating. Final resuspended volume of cells prior to plating was 12.8mL. Cells were plated (200uL/plate) on TDO plates (150mm) and incubated 30C until Monday. Dilutions of 1:100, 1:1000, and 1:10,000 were plated onto SD/-LT (200uL/plate; 100mm) to assess mating efficiency. Colonies from TDO plates will be restreaked onto QDO plates on Monday.PCR - Y2H Colonies: Co-trans MSTN1b active + O.mykiss red muscle cDNA library for sequencing
This is being performed on colonies from 20081027. PCR is being run with Immomix and plate will be sent directly to sequencing facility. See Sequencing Log for plate layout/primers. PCR set up here.2D Gel - V.tubi anoxia, gigas gill samples CONTINUED (from yesterday)
IEF was performed on the 4 strips from yesterday with the following parameters:
250V - 15 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
Dye fronts on the strips did not migrate at the same rates, even within experimental sets. This is a bit disconcerting, as these should all hae the same amount of protein AND they have all been desalted.
Strips were incubated with 10mL of sample reducing buffer for 15mins. w/shaking and then 10mL of sample alkylation buffer for 15mins. w/shaking.
Strips were removed and loaded into Invitrogen ZOOM 4-20% tris-glycine gels with the '+' end of the trips closest to the ladder. Strips were covered with 400uL 0.5% agarose (made with 1X tris-glycine running buffer) . Agarose was allowed to solidify for 10mins. 10uL of SeeBlue ladder was loaded into ladder lane.Gels were run 125V for ~2hrs.
Gels were stained according to Invitrogen SilverQuest protocol.
Gigas U1. Unedited picture here.

Gigas T1. Unedited picture here.
Vt aerobic 6hr. Unedited picture here.
Vt anoxic 6hr. Unedited picture here.

Results: The gigas gels look EXTREMELY overloaded. Not sure why, since the absorbance values obtained during quantitation were VERY consistent, suggesting that the assay was good. The Vt gels also look slightly overloaded, particularly in proteins of higher molecular weight. Steven has suggested running gels with half of what was used today in order to simply establish a nice looking reference gel.
20081030
2D Gel - IPG Strip Rehydration: V.tubi anoxia samples from 20081023, gigas gill samples from today
See Tatyana's notebook for volumes/gel on 20081027. Used half the volumes she loaded on that gel. Will load 15ug protein from each sample onto each strip (Invitrogen IPG 3-10NL).| SAMPLE |
Volume for 15ug (uL) |
Water to reach Vf = 155uL (uL) |
| U1 |
4.94 |
7.46 |
| T1 |
4.92 |
7.48 |
| Anoxia 6hr. |
11 |
1.4 |
| Aerobic 6hr. |
8 |
4.4 |
Protein Isolation/Prep - Gigas V.tubi 24hr exposure (from 20081008 & 20081009)
0.25g of gill from oysters U1 and T1 were resuspended in 0.5mL of Mammalian CellLytic + protease inhibitor cocktail and ground with a disposable pestle. An additional 0.5mlwas added to each sample and mixed. Tubes were spun 13,000g 10mins 4C. Supe was trnasferred to new tube and spun 13,000g 10mins. 4C. Supe was transferred to YM-3 (3kDa NMWL) Centricon filter and spun 7000g 1hr 4C. 1mL of water was added to each Centricon after this first spin and spun for an additional 1hr under the same conditions for desalting.Volume hasn't changed. Continued spinning for an additional 1hr. Still hasn't changed. Column probably clogged. Transferred retentate to new YM-3 column. Spun 7000g 15mins. 4C. Liquid passed through filter. Transferred retentate to 1.5mL snap cap tube.
Protein quantitation was done using the Pierce Bradford Coomassie reagent by mixing 5uL of each sample with 250uL of RT reagent. Mixed well and incubated @ RT for 10mins. Measured OD595 on NanoDrop.
Concentrations were detemined using the following equation:
[sample] = (Avg. OD595 - 0.04 + 0.0403)/0.00007
Equation was previously determined from a set of BSA standards.
| OD595 |
Avg. OD595 |
[Protein] (ug/mL) |
|
| U1 |
0.216 |
||
| 0.215 |
|||
| 0.209 |
|||
| 0.210 |
|||
| 0.212 |
0.2124 |
3038.57 |
|
| T1 |
0.222 |
||
| 0.215 |
|||
| 0.217 |
|||
| 0.202 |
|||
| 0.210 |
0.2132 |
3050 |
pBAD Expression - Dialysis of Decorin, FST, LAP & Telethonin CONTINUED (from yesterday)
1x TBS buffer was changed twice: once in the morning and again in the afternoon. Will continue incubation O/N with stirring @ 4C.Y2H: Trout muscle cell library mating CONTINUED (from yesterday)
OD600 of Y187[[pBAD/MSTN1b active] at 40hrs was 1.125. This is good. Cells were pelleted 600g , 5mins, 25C and then resuspended in 5mL SD/-T. A 1mL aliquot of AH109[trout muscle cell culture library] was thawed in a RT water bath. These two cultures were combined and added to a sterile 2L flask containing 45mL 1x YPDA + Kan50. Culture will be incubated 30C, 40RPM for 24 hrs and then plated on large TDO plates.
20081029
pBAD Expression - Dialysis of Decorin, FST, LAP & Telethonin CONTINUED (from 20081024 & 20081027)
Resuspended bacterial pellets in pBAD Lysis Buffer. Used 10mL for Decorin and FST, since these were 1L cultures to start with. Used 5mL for LAP & Telethonin since these were 500mL cultures to start. Sonicated cell suspensions @ 50% power, using 5 x 20 second pulses. Suspensions were pelleted 4000RPM, 4C, 45 mins. Supe (soluble proteins) was transferred to Fisher dialysis tubing (NMWL 12-14kDa). Pellets were resuspended in 4X Binding/Solubilization buffer. Suspension was spun 4000RPM, 4C, 30mins. Supe was transferred to Fisher dialysis tubing (NMWL 12-14kDa). All dialysis tubes were put into 4L of 1x TBS and incubated with stirring O/N @ 4C.Y2H: Trout muscle cell library CONTINUED (from yesterday)
OD600 of Y187[pBAD/MSTN1b active] at 24hrs inucbation was 0.0462. Need OD600 to >0.8. Will incubate O/N again to allow culture to reach proper density.
Protein Digestion - Bands from MSTN1b + red muscle extract Co-IP CONTINUED (from yesterday)
I screwed this up. Don't want to risk it. Will redo muscle extraction and co-ip. :(20081028
Protein Digestion - Bands from MSTN1b + red muscle extract Co-IP (from 20081022)
Bands were destained according to Invitrogen SilverQuest protocol. Digestion was done according to Goodlett Lab protcool. Incubated bands O/N, according to protocol. Will finish processing them tomorrow and submit one for sequencing.Y2H: Trout muscle cell culture library
Initial SD/-L plates from 20081021 were chilled @ 4C for 4hrs. Colonies were collected from plates by using 10mL of sterile Freezing Medium (1x YPDA + 25% glycerol) and sterile glass beads for every 2 plates with gentle swirling. Medium was removed from plates and temporarily stored in a sterile flask. Accumulated yeast was then divided into 1mL aliquots and stored @ -80C.To prepare for mating, inoculated 50mL of SD/-L + Kan20 with Y187[pBAD/MSTN1b active] in 250mL flask. Incubated @ 30C, 250RPM O/N.
pBAD Expression - Insoluble/soluble check for Decorin, FST, LAP, Telethonin CONTINUED (from yesterday)
Followed Western protocol for the remainder. Developed with Millipore Immobilon Chemiluminescent reagents.10min. exposure. Raw image. Light image.

Results: Positive control is visible (darkest band on membrane). Other bands visible on membrane likely belong to LAP. The failure of this experiment is likely related to the incubation with the primary Ab. When I arrived the following morning, the plastic-wrapped container was not level and it's possible that the solution did not cover the entire membrane.
20081027
pBAD Expression Western - Insoluble/soluble check for Decorin, FST, LAP, Telethonin
Will check the above proteins to determine their solubility according to pBAD protocol. Samples were loaded onto a 4-20% Pierce Tris-Hepes SDS/PAGE gel, along with 10uL SeeBlue ladder and 5uL of positive control lysate. Gel was run 150V for 40 mins. Transferred to nitrocellulose membrane (equilibrated in tris-glycine transfer buffer) 15V, 15mins. Gel was Coomassie stained for 1hr and then destained in 10% acetic acid until bands were clearly visible. The membrane was blocked 1hr. Added previously used (used thrice before on 20081024, 20081015, 20081023; stored in blocking solution, 4C) primary Ab (anti-6x His). Incubated O/N @ 4C.pBAD Expression: LAP, Telethonin CONTINUED (from yesterday)
Inoculated 500mL LB Amp100 with 5mL of O/N cultures. Incubated 3hrs 37C 230RPM. Induced expression of LAP with 500uL of 20% arabinose (Cf = 0.02%) and induced expression of Telethonin with 5mL of 20% arabinose (Cf = 0.2%). Incubated 37C 230RPM 4hrs. Pelleted bacteria 4000RPM, 15mins., 4C. Removed supe and stored bottles @ -20C.Y2H: O.mykiss Red Muscle + MSTN1b active Co-Trans
Colonies from the initial TDO plates were restreaked onto QDO plates by Leslie, Rachel, and Tatyana, and incubated @ 30C to assess growth.20081026
pBAD Expression: LAP, Telethonin
Inoculated 15mL LB Amp100 with LAP colony #5 and Telethonin colony #3 in 50mL conicals. Incubated O/N 37C, 230RPM.20081024
SDS/PAGE - pBAD Expression: LAP, Telethonin
Using induction samples (1-5) prepped on 20081015. Boiled samples for 5mins and loaded 5uL of each LAP sample and 10uL of each Telethonin sample, 5uL of control lysate and 10uL of SeeBlue ladder onto Pierce 4-20% Tris-Hepes gel. Ran 150V, 45mins. Transferred to nitrocellulose membrane (equilibrated in tris-glycine transfer buffer) 15V, 15mins. Gel was Coomassie stained for 1hr and then destained in 10% acetic acid until bands were clearly visible. The membrane was blocked 1hr. Added previously used (used twice before on 20081015 and 20081023; stored in blocking solution, 4C) primary Ab (anti-6x His). Incubated 1hr. Followed Western protocol for the remainder. Developed with Millipore Immobilon Chemiluminescent reagents.10 minute exposure.
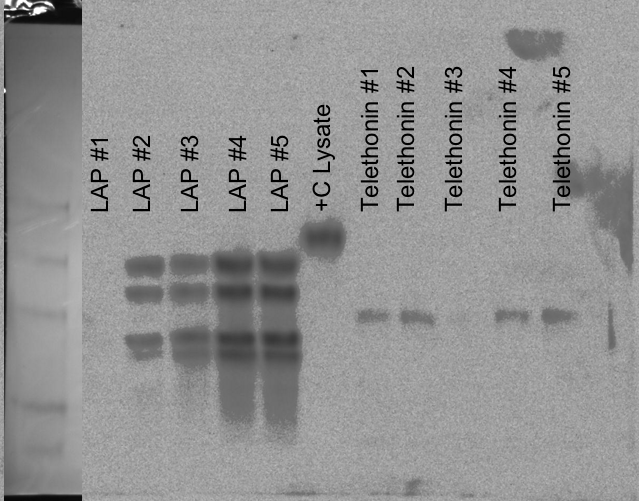
Results: LAP exhibits multiple bands; not sure why. Highest levels of expression are in LAP #4. Telethonin is WEAKLY expressed. Highest levels of expression are in Telethonin #5. Will inoculate starter cultures on Sunday O/N and induce expression with the proper amount of arabinose in larger cultures on Monday.
pBAD Expression: Decorin, FST CONTINUED (from yesterday)
Used 10mL of O/N cultures to inoculate 1L LB Amp100. Incubated 37C 230RPM for 3hrs. Induced expression in Decorin with 1mL 20% arabinose (Cf = 0.02%) and in FST with 100uL 20% arabinose (Cf = 0.002%). Incubated 4hrs 37C 230RPM. Cultures were pelleted 4000RPM 4C 15mins. Supe removed and bottles stored @ -20C.20081023
pBAD Expression: Decorin, FST
Inoculated 15mL LB Amp100 with Decorin colony #1 and FST colony #3 in 50mL conicals. Incubated O/N 37C, 230RPM.SDS/PAGE - pBAD Expression: Decorin, FST
Using induction samples (1-5) prepped on 20081015. Boiled samples for 5mins and loaded 10uL of each Decorin sample and 5uL of each FST sample, 5uL of control lysate and 10uL of SeeBlue ladder onto Pierce 4-20% Tris-Hepes gel. Ran 150V, 45mins. Transferred to nitrocellulose membrane (equilibrated in tris-glycine transfer buffer) 15V, 15mins. Gel was Coomassie stained for 1hr and then destained in 10% acetic acid until bands were clearly visible. The membrane was blocked 1hr. Added previously used (used once on 20081015; stored in blocking solution, 4C) primary Ab (anti-6x His). Incubated 1hr. Followed Western protocol for the remainder. Developed with Millipore Immobilon Chemiluminescent reagents.10 minute exposure.
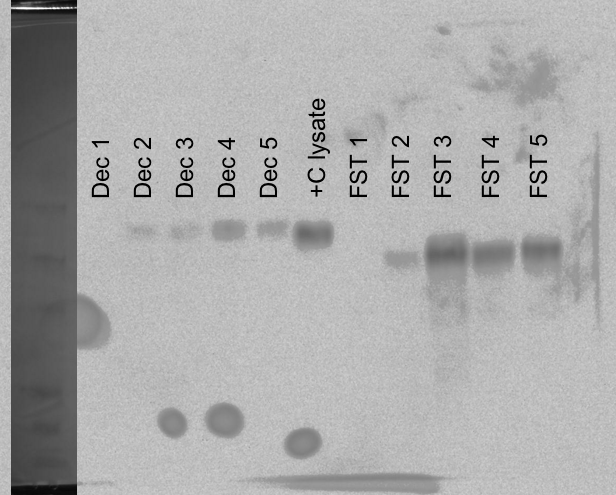
Results: The Decorin #4 sample shows the highest level of expression and the FST #3 sample shows the highest level of expression. Will inoculate starter cultures O/N and induce tomorrow with the appropriate amount of arabinose.
20081022
SDS/PAGE - MSTN1b Active/O.mykiss Red Muscle extract Co-IP
Co-IPs from yesterday were washed/eluted according to the Pierce anti-myc Co-IP Kit protocol. For each flow-through (>50kDa and <50kDa samples), 15uL was tranferred to fresh tube and mixed with and equal volume of 2x reducing sample buffer. To the elution samples, 2uL of 2-ME was added to each. All four samples were boiled for 5mins and then loaded onto a Pierce 4-20% tris-hepes gel. 10uL of SeeBlue ladder was loaded. Gel was run @ 150V for 40mins. Gel was removed and silver stained according to Invitrogen basic protocol.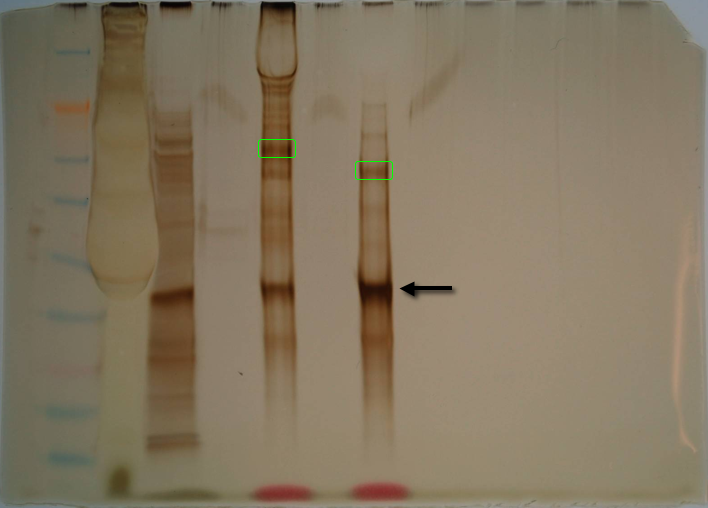
Lane 1 = Ladder
Lane 2 = >50kDa flow-through
Lane 3 = <50kDa flow-through
Lane 4 = empty
Lane 5 = >50kDa elution
Lane 6 = empty
Lane 7 = <50kDa elution
Results: The darkest band on the gel is MSTN1b active form and is indicated by the arrow. The two bands marked with the green boxes were excised and will be prepped for mass spec analysis.
20081021
2D Gels - V. tubi anoxic 6hr vs. aerobic 6hr.
See Tatyana's notebook for info.
Y2H Libraries - Trout muscle cells and O.mykiss red muscle
3mL 1x YPDA was inoculated in a glass culture tube with a fresh colony of AH109 and incubated 8hrs 30C, 200RPM. 5uL of this was used to inoculate 2 x 50mL 1x YPDA in 250mL flasks. Incubated 19 hrs. 30C, 230RPM. Cells were pelleted 700g, 5mins and resuspended in 100mL 1x YPDA. Incubated 4 hrs. 230RPM, 30C; OD600 = 0.8375.Competent AH109 cells were prepared according to Clontech Protocol. Transformations were done according to Clontech protocol. Co-transformation with red muscle was done with (10uL = 4.7ug) MSTN1b Mature M.P. DNA from DATE. Muscle cell library was plated on ~200 SD/-L plates. Co-trans was plated on ~40 TDO plates. Plates were incubated @ 30C.
20081020
Protein Expression - SDS/PAGE-Western Blot - Control, BMP, Decorin, FST, FST-L, LAP, MSTN
This is a repeat of that run on 20081015, but with brand new (i.e. not 2 years old) developer.Samples were boiled 5 mins. 10uL of each sample was loaded onto a Pierce 4-20% Tris-Glycine SDS/PAGE gel. 10uL of SeeBlue ladder was loaded. 5uL of positive control lysate was loaded in the same lane with the ladder. Gel was run @ 150V for 45mins.
Gel was transferred to nitrocellulose membrane (equilibrated in tris-glycine transfer buffer for 5mins) @15V for 15mins. Membrane washed briefly in 1x TBS-T, then blocked for 1hr. Gel was stained for 1hr in Coomassie stain and then destained w/ 10% acetic acid until bands were clearly visible.
Primary Ab (anti-6x His) was added to membrane at a 1:15,000 dilution. This incubated for 1hr.
Membrane was washed with 1x TBS-T and fresh blocking solution was added per the protocol.
Secondary Ab (DAR HRP) was added to membrane @ 1:15,000 dilution and incubated for 30mins.
Membrane was washed with 1x TBS-T per the protocol.
Western was developed using Millipore Immobilon Western Chemiluminescent reagent (2mL of each reagent) and imaged.
15 minute exposure.
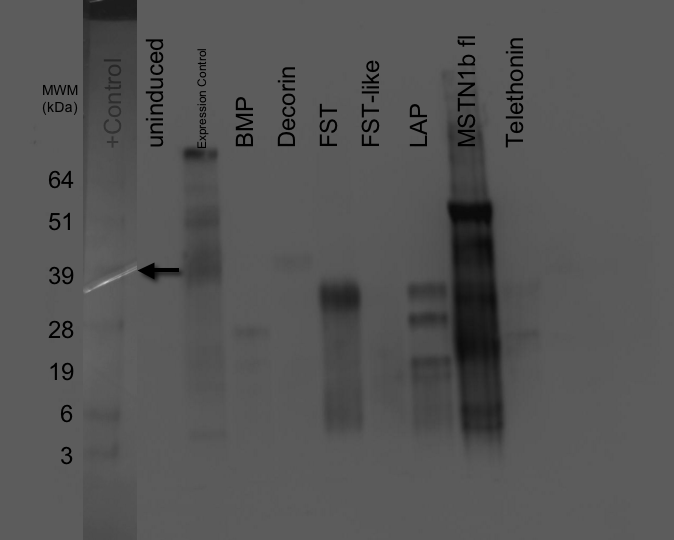
Results: Good expression from BMP, Decorin and FST. FST-like doesn't appear to have any detectable levels of protein. May need a different induction sample (e.g. 1,2,4, or 5). LAP shows mulitple bands and could be due to dimerization or cleavage. MSTN1b fl shows a strong, high-molecular weight band which could be correct. Also shows a multitude of other bands. Telethonin appears to show multiple bands, as such different induction samples should probably be examined.
The arrow on the blot indicates the position of the positive control, which runs @ 41kDa. The band is beneath the ladder overlay.
20081018
Gigas Hemocytes and V.tubiashii for challenges CONTINUED (from yesterday)
PGN-SA treated hemos were collected @ 24hr post-treatment. Water was removed and 1mL of Tri-Reagent was used to collect hemocytes from plates. Tubes were stored @ -80C in "Gigas Hemocytes" box.Protein Concentration/Desalting - V.tubiashii samples collected yesterday
One set of bacterial samples were resuspended in 2mL pBAD Lysis Buffer and subjected to 3 rounds of freeze/thaw cycles in dry ice and 42C water bath. Samples were transferred to a YM-3 Centricon spin column (cutoff = 3kDa) and spun @ 6000g, 2hrs., 25C. The remaining liquid on top of the filter was transferred to a clean tube and stored @ -20C.20081017
Gigas Hemocytes and V.tubiashii for challenges CONTINUED (from yesterday)
The water was removed from the plates. The plates were GENTLY washed with 5mL cold sea water. 5mL of sea water was then added to 3 of the 5 plates. Two of these three plates each received 15ug (3ug/mL) PGN-SA. The remaining two plates each received 3.5 mL of the V.tubi cultures (OD600 = 1.51). All plates were put back in the incubator.PGN-SA plates were incubated for 4 hr and 24hr. At those points, water was removed and 1mL of Tri-Reagent was used to collect hemocytes from plates. Tubes were stored @ -80C in "Gigas Hemocytes" box.
V.tubiashii plates: Liquid was collected from the plates, split into two equal volumes and pelleted @ 400RPM, 15mins, 4C. Supernatant removed and tubes stored @ -80C. After removal of liquid, plates were washed with 1mL Tri-Reagent. Tubes were stored @ -80C.
V.tubiashii for anoxia experiment CONTINUED (from yesterday)
Only one of the two cultures had grown. This culture was split into 4 X 10mL into sterile 250mL flasks. Volumes were adjusted to 100mL with LB+1%NaCl. OD600=0.306. Attempted to vacuum-seal a culture to eliminate O2/induce anoxic conditions. This failed. Mac had idea to simply fill two 50mL concials all the way to the top to eliminate any headspace. These two tubes and two flasks were incubated @ ~30C (as cool as Graham's inubator would get), 200RPM. Cultures were collected at 3 and 6hrs. Each culture was split into two tubes: 1 for protein sample and 1 for RNA sample. Cells were pelleted by spinning 15 mins. 4000RPM, 4C. Supe was discarded and tubes were stored @ -80C.| Culture |
OD600 @ 0hrs |
OD600 @ 3hrs |
OD600 @ 6hrs. |
| Aerobic |
0.306 |
1.4 |
1.54 |
| Anoxic |
0.306 |
0.63 |
0.665 |
20081016
Gigas Hemocytes and V.tubiashii for challenges
Withdrew ~20mL hemocytes from 8 C.gigas (2 large, 6 small). Hemocytes were stored on ice, spun 100xg, 15mins, 4C. Supe was removed and hemocytes were resuspended in 20mL cold, sterile sea water + Pen/Strep (1:100 dilution). Hemos were plated on poly-d-lysine plates (4 plates x 5mL/plate) and incubated O/N @ 12C in the dark.V.tubiashii colonies were used to inoculate 5mL of LB+1%NaCl (x 4) in 50mL conicals. Cultures were incubated @ ~30C (as cool as Graham's inubator would get), 200RPM O/N.
V.tubiashii for anoxia experiment
Inoculated 50mL LB+1%NaCl (x2) in sterile 250mL flasks. Cultures were incubated @ ~30C (as cool as Graham's inubator would get), 200RPM O/N.20081015
Protein Expression - SDS/PAGE-Western Blot - Control, BMP, Decorin, FST, FST-L, LAP, MSTN
Samples were boiled 5 mins. 5uL of each sample was loaded onto a Pierce 4-20% Tris-Glycine SDS/PAGE gel. 10uL of SeeBlue ladder was loaded. 2uL of positive control lysate was loaded in the same lane with the ladder. Gel was run @ 150V for 45mins.Gel was transferred to nitrocellulose membrane (equilibrated in tris-glycine transfer buffer for 5mins) @15V for 15mins. Membrane washed briefly in 1x TBS-T, then blocked for 1hr. Gel was stained for 1hr in Coomassie stain and then destained w/ 10% acetic acid until bands were clearly visible.
Primary Ab (anti-6x His) was added to membrane at a 1:15,000 dilution. This incubated for 1hr. Ab solution was stored @ 4C.
Membrane was washed with 1x TBS-T and fresh blocking solution was added per the protocol.
Secondary Ab (DAR HRP) was added to membrane @ 1:15,000 dilution and incubated for 30mins.
Membrane was washed with 1x TBS-T per the protocol.
Western was developed using ECL Plus (2mL of reagent A and 50uL of reagent B) and imaged.
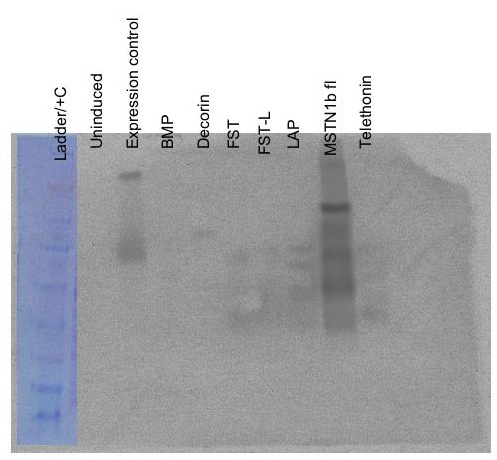
20081014
Protein Expression - SDS/PAGE Decorin, FST, FST-L, MSTN
Pellets were resuspended in 80uL of 1x reducing sample buffer and boiled for 5mins. 5uL of each sample was loaded onto a Pierce 4-20% SDS/PAGE gel and run @ 150V for 45 mins. 10uL of SeeBlue molecular weight marker was loaded onto each of the two gels. Gels were stained for 30 mins. with Coomasie and destained with 10% acetic acid until bands were clearly visible.Gel #1 ------------------------------------------------------------ Gel #2
Gel #1
Lane 1 = SeeBlue ladder
Lane 2 = Pre-induced MSTN
Lanes 3-5 = Decorin 1-5
Lanes 6-12 = FST 1-5
Gel #2
Lane 1 = Pre-induced MSTN
Lanes 2-6 = FST-L 1-5
Lane 7 = SeeBlue ladder
Lanes 8-12 = MSTN1bfl 1-5
Y2H Libraries - Red muscle, white muscle, muscle cells
mRNA isolation CONTINUED (from yesterday)
mRNA was precipitated. White muscle and cell culture mRNA were resuspended in 5uL and red muscle mRNA in 10uL of "The RNA Storage Solution". Samples were spec'd. 260/280 ratios for all three samples are rather low, but 26/280 ratios look OK. Yield for muscle cells is still extraordinarily high (nearly 1ug).First strand cDNA synthesis
Used 3uL of mRNA from each sample. Followed Clontech protocol. At the end of this porition of the protocol, transferred 2uL of rxn to new, chilled tube and stored rest of first-strand rxn @ -20C.
Amplify ds cDNA by LD-PCR
Performed PCR according to Clontech protocol using 2uL from First strand cDNA synthesis. Ran 20 cycles. After completion of PCR, rxn was stored @ -20C.Protein Expression - BMP CONTINUED (from yesterday)
Inoculated 6 (1-5 & control sample) 50 mL conicals containing 10mL LB+Amp100 with 100uL of culture started yesterday. Incubated 3hrs 37C, 200RPM. Took 1mL (x2) of control samples from each, pelleted, stored @ -20C. Added appropriate amounts of arabinose to appropriate tubes. Incubaed 4hrs 37C, 200RPM. Took 1mL (x2) from each tube, pelleted, stored @ -20C.20081013
Protein Expression - BMP
Colony #2 (see gel from yesterday) was used to inoculate 5mL of LB+Amp100 in a glass culture tube. Incubated O/N 37C, 200RPM.Y2H Libraries - mRNA isolation: O.mykiss red/white muscles, Steven's muscle cell cultures
Total RNA from these samples was precipitated O/N (see yesterday). mRNA was isolated according to Ambion protocol. mRNA was quantified. Yields for both muscle mRNAs look reasonable (~1ug total), while the yeild from the muscle cell culture mRNA seems extraordinarily high (~1ug). To improve accuracy of quantitatino and reduce the sample volume, the mRNAs were preciptated according to the Ambion procol. These were stored O/N @ -20C.Protein Expression - FST, LAP, Control vector, MSTN1b CONTINUED (from yesterday)
Inoculated 6 (1-5 & control samples) 50 mL conicals containing 10mL LB+appropriate antibiotic with 100uL of cultures started yesterday. Incubated 3hrs 37C, 200RPM. Took 1mL (x2) of control samples from each, pelleted, stored @ -20C. Added appropriate amounts of arabinose to appropriate tubes. Incubaed 4hrs 37C, 200RPM. Took 1mL (x2) from each tube, pelleted, stored @ -20C.20081012
Y2H Libraries - RNA Precipitation: O.mykiss red & white muscle, Steven's muscle cell cultures
Used 20ug (20uL) of total RNA from red & white muscle RNA (from 20080104). Added 2uL 5M ammonium acetate (0.1 volumes) and 55uL 100% EtOH (2.5 volumes). Vortexed and incubated @ -20C O/N.Pooled total RNA from select muscle cell culture total RNA (See workup sheet. [RNA] were taken from Steven's notebook #8, 6/24/08.). Total RNA was 7.25ug in 22uL. Added 2.2uL 5M ammonium acetate (0.1 volumes) and 60.5uL 100% EtOH (2.5 volumes). Vortexed and incubated @ -20C O/N.
Protein Expression - FST, LAP, Control vector, MSTN1b
Inoculated 5mL LB+Amp100 with FST colony #3 and LAP colony #5 in glass culture tubes. Incubated 37C, 250 RPM O/N. Also inoculated 5mL LB+Amp100 with Decorin (colony #1), FST-L (colony #1), Telethonin (colony #3) and the control expression vector colony (will be used for induction) to grow up for mini-prep and frozen stocks. Also inoculated 5mL LB+Kan50 with pBAD/MSTN1bfl as a control for protein induction.PCR - Vector/Internal primers: BMP
Checking restreaked colonies using new pBADTA-F primer (Tm of 48.2) and gene-specific reverse primers. PCR will run O/N. PCR set up is the same as 20081006. If this new primer works, this PCR will show the orientation of the inserts in the vector.
Gel was loaded with 100bp Ladder in Lane #1, BMP colonies #1-10, and a negative control, in this order from left to right.
Results: All colonies appear to have produced the correct size band, but some colonies have a greater amount of PCR product. Will select one of those colonies for protein expression.
20081011
PCR - Vector/Internal primers: Decorin, FST, FST-L, LAP, & Telethonin
Checking restreaked colonies using new pBADTA_F primer (Tm of 48.2) and BMP_pBADTA_R primers. PCR will run O/N. PCR set up is the same as 20081006. If this new primer works, this PCR will show the orientation of the inserts in the vector.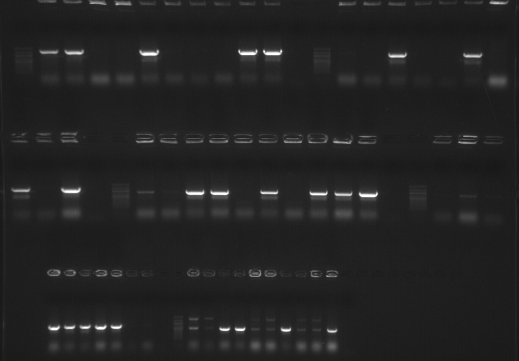
The gel is loaded in the order that the genes are listed in the table from yesterday, colonies 1-10 (from left to right for each gene) and then a negative control. 100bp Ladder separates genes.
Results: The colonies originally selected for protein expression for FST and LAP (both colonies #1) did not produce a band. Will inoculate cultures using a colony that produced band for protein expression. Decorin (colony #1), FST-L (colony #1) and Telethonin (colony #3) all show a band of appropriate size. Unfortunately, the lack of protein induction (see gels from yesterday) cannot be explained. Will repeat the SDS/PAGE gels using the secondary samples, transferred to screw cap tubes before boiling and load only 5uL. Hopefully this will help produce cleaner gels with more discrete banding.
Subcloning - BMP into pBAD CONTINUED (from yesterday)
Colonies grew. 10 colonies were restreaked onto gridded LB+Amp100 plate. Incubated O/N 37C.20081010
Subcloning - BMP into pBAD
Purified BMP PCR product from earlier today was cloned into pBAD following pBAD TOPO TA protocol. 1uL of purified PCR product was used. Reaction was incubated for 5 mins. TOP10 cells were transformed with 2uL of ligation reaction according to pBAD TOPO TA protocol. 50uL were plated on LB+Amp100 plates and incubated O/N, 37C.Protein Expression - Decorin, LAP & Telethonin CONTINUED (from yesterday)
80uL of 1x sample loading buffer (reducing) was added to each pellet, mixed and boiled for 5mins. 10uL of SeeBlue ladder were loaded. 10uL of each sample wer loaded onto a Pierce 4-20% SDS/PAGE gel in 1x Tris-Hepes running buffer and run @ 150V for 45 mins. Gels were stained for 30 mins. in Coomassie blue and then destained with 10% acetic acid until bands became clearly visible.| Protein |
#aa |
Molecular Weight (kDa) |
Expected Mol Weight Band on Gel(kDa) |
| Decorin |
364 |
40.4 |
42.4 |
| LAP |
238 |
26.6 |
28.6 |
| Telethonin |
184 |
21.4 |
23.4 |
Gel #1
Lane 1 = Ladder
Lane 2 = Uninduced
Lanes 3 - 7 = Decorin induction
Lanes 8 - 12 = LAP induction
Gel #2
Lane 1 = Ladder
Lane 2 = Uninduced
Lanes 3 - 7 = Telethonin induction
Results: Well, Gel #1 looks horrible. Likely due to water entering sample tubes during boiling. Protein load is high for both gels. However, there does not appear to be induction of anything! Will run PCR using new pBADTA_F primer and gene specific reverse primer on colonies to determine orientation of inserts.
PCR - BMP for pBAD cloning/expression
Primers = BMP_CDS_F, BMP_pBADTA_R. Template = TCAY0015H13. PCR set up here. Expected size = 774 bp.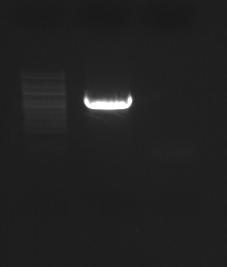
Results: PCR produced band of expected size. Will excise from gel and purify using Millipore spin columns.
Protein Expression - FST, FST-L CONTINUED (from yesterday)
100uL of each O/N culture was used to inoculate 6 x 10mL of LB+Amp100. These cultures were grown for 3 hrs (OD600 = 0.51) @ 37C, 250 RPM. A 1mL sample was taken from each control sample, pelleted and frozen @ -20C. Protein induction was performed by adding varying concentrations of arabinose to the appropriate cultures, as described in the pBAD TOPO TA protocol, and incubated 37C 250 RPM for 4 hrs. 1mL aliquots (x2/culture) were collected, pelleted and frozen @ -20C.20081009
Protein - Expression - FST, FST-L
Inoculated 2 x 5mL LB+Amp100 in glass culture tubes with FST colony #1 and FST-L colony #1 for protein expression. Incubated O/N 37C, 250RPM.Protein Expression - Decorin, LAP & Telethonin CONTINUED (from yesterday)
100uL of each O/N culture was used to inoculate 6 x 10mL of LB+Amp100. These cultures were grown for 3 hrs (OD600 = 0.51) @ 37C, 250 RPM. A 1mL sample was taken from each control sample, pelleted and frozen @ -20C. Then, protein induction was performed by adding varying concentrations of arabinose to the appropriate cultures, as described in the pBAD TOPO TA protocol , and incubated 37C 250 RPM for 4 hrs. 1mL aliquots (x2/culture) were collected, pelleted and frozen @ -20C.20081008
Protein Expression - Decorin, LAP & Telethonin
In an effort to speed things along, inoculated 3 x 5mL LB+Amp100 in glass culture tubes with Decorin colony #1, LAP colony #1, & Telethonin colony #3 for protein expression before finding out the result of the PCR from today. Incubated O/N 37C, 250RPM.Subcloning - Genes for MSTN Project CONTINUED (from today)
Due to difficulties with checking the resulting colonies from the cloning rxn (see the results of the gel below), I will rescreen the colonies using just gene-specific primers. Unfortunately, can't use vector primers because there are an insufficient amount included with the kit for checking colonies. Additionally, the sequence of the pBAD TOPO TA vector appears to differ from the pBAD202/TOPT that we used to use. As such, the existing forward primer we've used in the past for pBAD will not work. The reverse primer, however, remains the same.PCR set up is the same as 20081006 . PCR was run O/N.
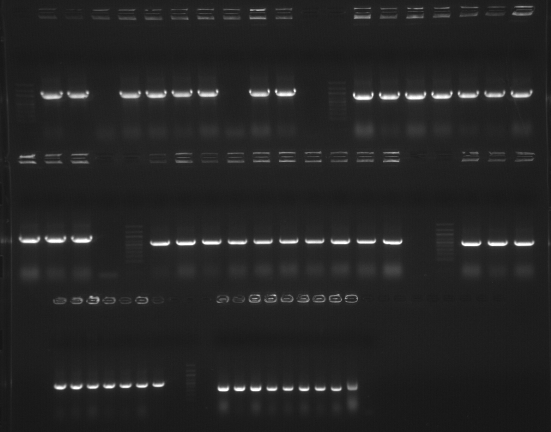 |
| external image 20081009.png |
The gel is loaded in the order that the genes are listed in the table from yesterday, colonies 1-10 (from left to right for each gene) and then a negative control. 100bp Ladder separates genes.
Results : Most of the colonies screened appear to have the appropriate insert. Correct orientation of the insert is still not determined, though. Orientation will be assessed with either a combo of the new pBADTA_F primer and an internal primer OR performing restriction digestions on miniprep DNA from candidate colonies.
Subcloning - Genes for MSTN Project CONTINUED (from 20081007)
Streaked colonies were re-screened in the same manner as yesterday . Hopefully this will produce better results and lead to identification of more positive clones.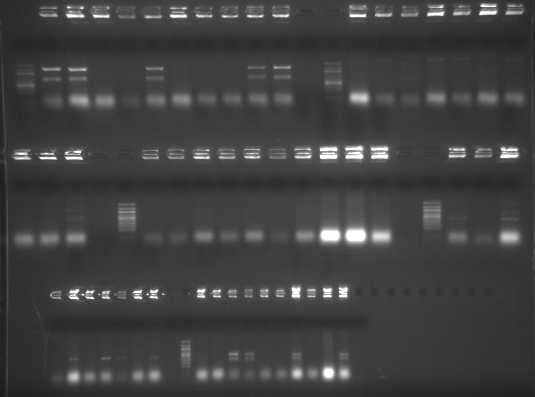 |
| external image 20081008.png |
The gel is loaded in the order that the genes are listed in the table from yesterday, colonies 1-10 (from left to right for each gene) and then a negative control. 100bp Ladder separates genes.
Results : In essence, these are the same results I got yesterday, but brighter bands. Every "working" rxn shows multiple bands. Genes that show a band of the appropriate size are Decorin (colonies 1,2,5,9,10) and Telethonin (colonies 3,4,7,10).
Investigating this a bit futhers, it appears that the Tm for BAD_R primer is only 41.8C! Our gene specific primers are all around 50-55C. This difference in Tm could explain the questionable results. To simply determine the presence (or lackthereof) of our inserts, I'll screen the streaked colonies via PCR with just gene-specific primers. However, this will not provide us with any information regarding directionality of the inserts.
20081007
Gigas V.tubiashii Challenge CONTINUED (from yesterday)
Four oysters, two per container, were allowed to acclimate to RT O/N. 1L of Lb + 1%NaCl was incoluated with the 10mL culture started yesterday. This will incubate O/N @ 25C, 200RPM.Subcloning - Genes for MSTN Project CONTINUED (from 20081006)
Resulting colonies (10 from each cloning reaction) will be streaked onto a gridded plate and then PCR'd. PCRs will be conducted using a vector primer and a gene-specific primer to confirm successful ligation as well as correct orientation of the gene in the vector. PCR set up here .
| Gene |
Expected Product Size |
Primers |
| Decorin |
~1235 |
Decorin_CDS_F, pBAD_R |
| FST |
~1100 |
FST_CDS_F, pBAD_R |
| FST-L |
~900 |
FST-L_CDS_F, pBAD_R |
| LAP |
~850 |
LAP_CDS_F, pBAD_R |
| Telethonin |
~700 |
Telethonin_CDS_F, pBAD_R |
 |
| external image 20081007.png |
The gel is loaded in the order that the genes are listed in the table above, colonies 1-10 (from left to right for each gene) and then a negative control. 100bp Ladder separates genes.
Results : Well, there are bands in some colonies from Decorin and LAP. Colonies that were selected were rather small and possibly did not provide sufficient starting material for PCR after being streaked on the gridded plate? Will PCR restreaked colonies tomorrow to assess this possibility.
20081006
Gigas V.tubiashii Challenge
Inoculated 10mL LB + 1%NaCl with a V.tubiashii colony. Incubated O/N 25C, 200RPM.Subcloning - Genes for MSTN Project CONTINUED (from 20081004)
Bands cut from gel were purified using Millipore spin columns. Bands were cloned into pBAD via the pBAD TOPO TA kit using 1uL of PCR product from each purified band. The cloning reaction was run for 5 mins. 2uL of each reaction (including LAP from 20081002 ) were used to transform Top10 cells and 50uL of cells (remainder of cells stored @ 4C) were plated on LB/Amp100 plates and incubated O/N @ 37C.20081004
Subcloning - Genes for MSTN Project
Repeated the exact same PCR conditions from 20081002 using the templates/primers listed below. BMP and LAP were not included in this round because need new Reverse BMP primer (to eliminate stop codon) and LAP amplification from 20081002 is fine.| Gene |
Template |
Expected Product Size |
Primers |
| Decorin |
cDNA (R. Trout Red Muscle from 6/20/2007) |
1095 |
Decorin_CDS_F, Decorin -> pBAD |
| FST |
IRT70J17BE09 (EST clone) |
969 |
FST_CDS_F, FST -> pBAD |
| FST-L |
TCBK0035L03 (EST clone) |
759 |
FST-L_CDS_F, FST-L -> pBAD |
| Telethonin |
TCBA0008D04 |
555 |
Telethonin_CDS_F, Telethonin -> pBAD |
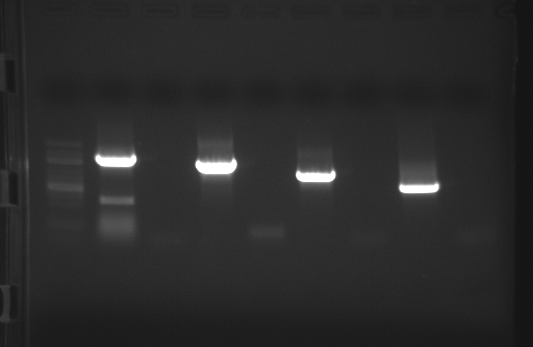 |
| external image 20081006.png |
Lane 2 - Decorin
Lane 3 - Decorin Neg. Control
Lane 4 - FST
Lane 5 - FST Neg. Control
Lane 6 - FST-L
Lane 7 - FST-L Neg. Control
Lane 8 - Telethonin
Lane 9 - Telethonin Neg. Control
Results : Gel looks good. The brightest band in each lane will be excised and purified.
Subcloning - Genes for MSTN Project CONTINUED (from yesterday)
Ugh. Realized wrong primers were used for PCR reactions. Primers used for all PCRs retained the stop codon for all the genes (except for LAP). Will redo PCRs/cloning.20081003
Subcloning - Genes for MSTN Project
Bands from the PCR run yesterday were excised and purified using Millipore spin columns. Bands were cloned into pBAD via the pBAD TOPO TA kit using 1uL of PCR product from each purified band. The cloning reaction was run for 5 mins. 2uL of each reaction were used to transform Top10 cells and 100uL of cells (remainder of cells stored @ 4C) were plated on LB/Amp50 plates and incubated O/N @ 37C.V.tubiashii pH Viability NEW CONTINUED (from 20081002)
After 18.5 hrs of growth, 1mL of each sample was removed and spec'd @ 595nm. After readings were taken, cultures were split evenly (4.5 mL and 4.55mL), spun @ 4000 RPM, 25C, 10 mins, supe was removed and samples were stored @ -80C.
Results :
| Sample |
OD595 |
| pH = 4 |
0.036 |
| pH = 5 |
0.041 |
| pH = 6 |
1.42 |
| Unaltered media (pH = 6.72) |
1.389 |
| pH = 7.13 |
1.348 |
| pH = 8 |
1.398 |
| pH = 9 |
1.447 |
| pH = 10 |
1.204 |
20081002
PCR - Genes for MSTN Project
Performed PCR O/N to get full length CDS for TA cloning into pBAD expression system on: BMP, Decorin, FST, FST-L, LAP, and Telethonin.| Gene |
Template |
Expected Product Size |
Primers |
| BMP |
TCAY00015BH13 (EST clone) |
774 |
BMP_CDS_F/R |
| Decorin |
cDNA (R. Trout Red Muscle from 6/20/2007) |
1095 |
Decorin_CDS_F/R |
| FST |
IRT70J17BE09 (EST clone) |
969 |
FST_CDS_F/R |
| FST-L |
TCBK0035L03 (EST clone) |
759 |
FST-L_CDS_F/R |
| LAP |
cDNA (R. Trout Red Muscle from 6/20/2007) |
714 |
LAP_CDS_F/R |
| Telethonin |
TCBA0008D04 |
555 |
Telethonin_CDS_F/R |
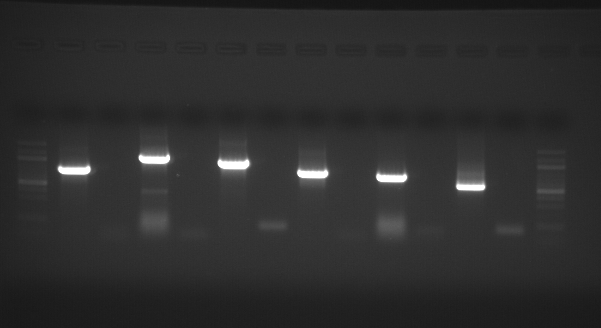 |
| external image 20081003.png |
Lane 1 = 100bp Ladder
Lane 2 = BMP
Lane 3 = BMP Neg. Contol
Lane 4 = Decorin
Lane 5 = Decorin Neg. Control
Lane 6 = FST
Lane 7 = FST Neg. Control
Lane 8 = FST-L
Lane 9 = FST-L Neg. Control
Lane 10 = LAP
Lane 11 = LAP Neg. Control
Lane 12 = Telethonin
Lane 13 = Telethonin Neg. Control
Lane 14 = 100bp Ladder
Results : All PCRs look good and have appropriately sized bands. Will excise and subclone using pBAD TOPO TA kit.
V.tubiashii pH Viability NEW CONTINUED (from 20081001)
Inoculated 8 tubes (pHs = 4, 5, 6, 6.72 (unadjusted media), 7, 8, 9, 10) filled with 10mL of 1x LB + 1% NaCl with 100uL of starter culture from yesterday. Starting OD595 of these 8 tubes = 0.021. Incubated O/N 25C 250 RPM. Will check OD595 of all cultures tomorrow to help evaluate growth/response to different pHs.20081001
V.tubiashii pH Viability NEW
Set up 50mL conicals containing 10mL 1x LB + 1% NaCl. The pH of each tube was adjusted and then the tubes were autoclaved. pHs = 4, 5, 6, 6.72 (unadjusted media), 7, 8, 9, 10. A 50mL conical tube containing 10mL 1x LB + 1% NaCl was inoculated with a V.tubiashii colony and incubated O/N 25C, 200RPM. Tomorrow will use 100uL of existing culture to inoculate each of the pHed media. Will incubate O/N 25C, 250RPM and will check OD595 the next day.V.tubiashii pH Viability CONTINUED (from 20080930)
All cultures have grown. Scrapped original idea/experiment. Changed experimental design to be easier/more controlled (See above).Estes Paper: http://www.int-res.com/abstracts/dao/v58/n2-3/p223-230/
20080930
V.tubiashii pH Viability
8 x 50mL conicals each containing 5mL of LB+1% NaCl were inoculated and incubated at 25C, 200 RPM O/N. Once culture growth is verified, the pH of each culture will be altered across a gradient and then culture viability will be assessed visually or spectorphotometrically.Gigas Hemocytes CONTINUED (from 20080929)
Pictures of the two plates were taken at 22 hrs. post-plating. RNA was isolated from both plates in a single prep by Mac. Total RNA yield was ~1ug.20080929
Gigas Hemocytes
~6mL hemocytes collected from 3 smaller oysters. Cells were diluted 1:1 with sterile sea water. 6mLs were plated on each of two poly D lysine plates and incubated at 15C O/N in the dark. Photographs were taken at 30 mins. and 3 hrs. post-plating and showed adherent cells. The 3 hr mark seemed to have a higher number of adherent cells than at the 30 min. time point.20080928
V.tubiashii + Gigas Hemocytes - Plates and liquid cultures CONTINUED (from 20080926)
Hemocytes cultures were bunk. No hemos! Experiment aborted. The liquid obtained from the oysters on 20080926 was likely just sea water.20080927
V.tubiashii + Gigas Hemocytes - Plates and liquid cultures CONTINUED (from 20080926)
V.tubiashii culture failed to grow. Will inoculate 5mL of MB in glass tube and two 50mL conical tubes with a large amount of V.tubiashii colonies and incubate O/N 37C, 200RPM. Hemocytes should be fine through tomorrow. This is based on past experience with hemocyte plating. However, hemocyte viability will be visually assessed tomorrow before adding V.tubiashii to plates.20080926
V.tubiashii + Gigas Hemocytes - Plates and liquid cultures.
~5mL of hemocytes were taken from 5 LARGE Pacific oysters. Very difficult to withdraw any liquid from muscles.Inoculated 5mL marine broth in glass culture tube with V.tubiashii colony. Incubated O/N @ 37C, 200 RPM.
Poly D Lysine Plates were incubated @ 18C O/N in the dark in order to allow the hemos to adhere to the plates. V.tubiashii will be added tomorrow.
Plate #1 = 1mL hemos + 5mL sterile SW (hemos only)
Plate #2 = 1mL hemos + 5mL sterile SW (hemos + Vt)
Plate #3 = 1mL hemos + 5mL sterile SW (hemos + MB)
Plate #4 = 6mL sterile SW (Vt only)
20080925
Real-time PCR - Vibrio fstz primers on undiluted cDNA.
Set up real-time PCR using undiluted cDNA with the ftsz primers.Sequencing Results from 20080919 are here.
User: samwhitePass: **ask***
Sequencing data is also available over at wet genes (no password needed!) -
| sr320 |
20080924
PCR - Test new Vibrio primers
PCR test of new Vibrio primers from yesterday yielded nothing, so I am retesting with undiluted/pooled from cDNA made 20080922 by Tatyana. Samples used for pooling were all Vt samples/time points. Used 2x Immomix . Primers tested:ftsz F/R
toxR F/R
PCR set up here. PCR ran over night.
@ sam : lets start scanning as images (not pdfs) -
| sr320 |
Did not retest ompW primers because a signal was obtained in the real-time PCR run yesterday.
Gel run 20080925
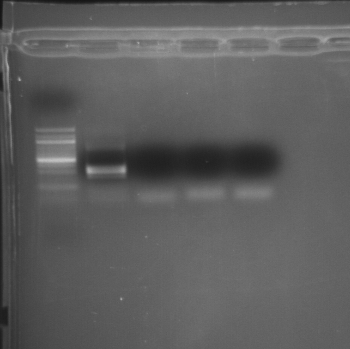 |
| external image 20080925-01.png |
Lane 1 = 100bp Ladder
Lane 2 = ftsz
Lane 3 = ftsz Neg. Control
Lane 4 = toxR
Lane 5 = toxR Neg. Control
Results: Undiluted cDNA pool produced multiple bands with the ftsz primers. Will run a real-time PCR using undiluted cDNA and the pooled cDNA as a positive control.
Real-time data analysis (from real-time 20080923)
This was performed using Real Time PCR Miner . Detailed analysis was performed according to protocol. Samples were normalized to 16s values. Fold over minimum was calculated. See how to perform Miner data analysis here. Data and graphs of this data are here.20080923
PCR - Test new Vibrio primers
Check new Vibrio primers:
ftsz F/R
ompW F/R
toxR F/R
Template: Diluted (1:4) cDNA from cDNA made yesterday by Tatyana ; V.t + Auto Gigas, t=0, 2, 4, 24hrs. Master mix, cycling params here.
Gel run 20080924.
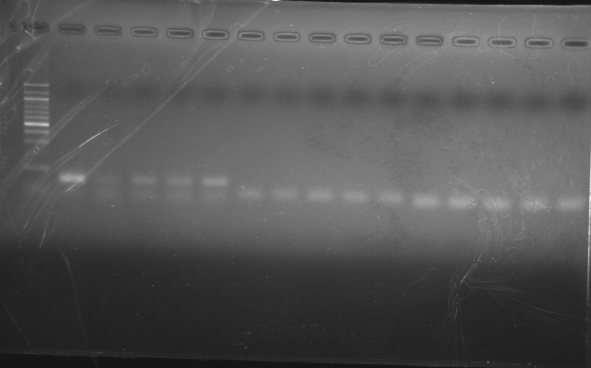 |
| external image 20080924.png |
Lane 1 = 100bp ladder
Lanes 2-6 = toxR t=0, 2, 4, 24, Neg. Control
Lanes 7-11 = ompW t=0, 2, 4, 24, Neg. Control
Lanes 12-16 = ftsz t=0, 2, 4, 24, Neg. Control
Real-time PCR - Vibrio Experiments Plate #2
Ran real-time PCR on diluted (1:4) cDNA made yesterday by Tatyana . Plate layout/master mix set up here .
Primers:
ompW F/R
ftsz F/R
Real-time PCR - Vibrio Experiments Plate #1
Ran real-time PCR on diluted (1:4) cDNA made yesterday by Tatyana . Plate layout/master mix set up here .
Primers:
V.tubi 16s F/R
V.tubi chitinase F/R
V.tubi VTHB F/R
toxR F/R
20080919
DNA Sequencing
Prepared sample and primer plates for sequencing submission. See Sequence Log for layout of sample/primer plates. Submitted 10uL of template for each sample and 3uM primer (3uL of 10uM primer + 7uL H2O per well) for each sample. Samples with less than 10uL were increased in volume with H2O to accommodate the number of wells for submission.
2D Gel - Desalted Vibrio protein samples (continued from yesterday)
IEF focusing was performed on the two strips from yesterday.
250V - 20 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
Incubated strips with 5mL reducing buffer, 15 mins. with shaking, then with 5mL alkylating buffer, 15 mins. with shaking.
Transferred strips to 4-20% ZOOM Tris-Glycine gels, '+' side nearest ladder. Loaded 10uL SeeBlue ladder in ladder well.
Ran 145 mins. @ 125V. Invitrogen rep says to run gel until dye front is at bottom of gel. Gels were stained according to Invitrogen Silverquest protocol.
V.tubi + H2O - Cut at upper right corner (above ladder)
 |
| external image 2871823533_b412272cf2.jpg |
V.tubi + gigas pool - Cut at upper left corner (opposite ladder)
 |
| external image 2871842483_a8529255a9.jpg |
Original Images
20080918
2D Gel - Desalted Vibrio protein samples (see Desalting section below)
Samples and Prep for IPG strip (ZOOM IPG pH 3-10 NL) rehydration O/N:
1. V.tubi + H2O - 12.65uL + 142.35uL sample rehydration buffer
2. V.tubi + gigas pool - 6.51uL + 148.49uL sample rehydration buffer.
RNA Isolation - Remaining Vibrio samples from 20080828
Isolated RNA with 1mL TriReagent from the following samples:
1. t=0, Vt + sterile sea water
2. t=0, Vt + live gigas
3. t=0, Vt + autoclaved gigas
4. t=0, gigas sea water
5. t=2, Vt + sterile sea water
6. t=2, Vt + live gigas
7. t=2, Vt + autoclaved gigas
8. t=2, gigas sea water
9. t=4, Vt + sterile sea water
10. t=4, Vt + live gigas
11. t=4, Vt + autoclaved gigas
12. t=4, gigas sea water
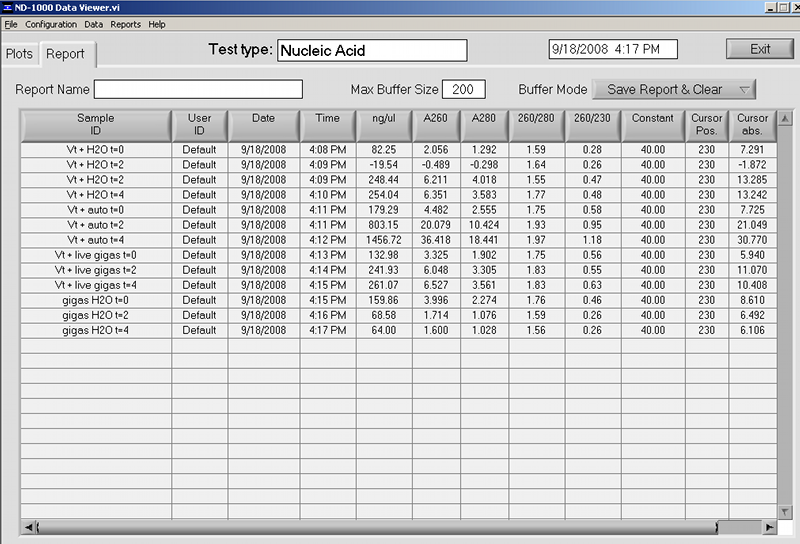 |
| external image Vt%20RNA%20SJW%2020080918.bmp |
Samples were stored in -80C, Shellfish RNA Box #3.
2D Gel - Gigas 24hr Vibrio Exposure, Vibrio 24hr gigas exposure Continued from 20080917
IEF focusing was performed on all 6 strips from yesterday.
250V - 20 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
NOTES: Strips 1 & 2 from yesterday both exhibited arching. Strip 2 showed signs of arching towards the end of the 750V step and strip 1 started arching at the beginning of the 2000V step. These strips were removed from the cassette when arching was detected. Both samples were extracted in pBAD Lysis Buffer which contains salts. Will put those preps through a Millipore Centricon 3kDa to remove salts.
After focusing, the cassette (containing samples 4-6 from yesterday) was stored in the -80C on the bottom shelf in a box labeled "2D IPG Strips".
Desalting/Concentration of Vibrio protein samples.
For Centricon will prep two samples -| sr320 |
- t=24hr w/ oysters & t=24hr w/autoclave oysters.
- t=24hr sterile water only
Spin 6000 g for 2hr
sample #2 had less protein as it was from one sampling tank as opposed to the sample #1 which was from two sampling tanks.
Checked after 2hr. Still about 300ul above, added 250ul water to each, spin another hour at 6g. -
| sr320 |
After total 3 hour spinning, took off ~200ul of sample from each -
| sr320 |
Samples were quantitated using the Pierce Coommassie Bradford assay .
Concentrations were detemined using the following equation:
[sample] = (Avg. OD595 - 0.04 + 0.0403)/0.00007
Equation was previously determined from a set of BSA standards.
| SAMPLE |
OD595 nm |
Avg. OD595 |
[sample] (ug/mL) |
| V.tubi + H2O |
0.084 |
||
| 0.086 |
|||
| 0.079 |
0.083 |
1186 |
|
| V.tubi + gigas pool |
0.147 |
||
| 0.169 |
|||
| 0.158 |
|||
| 0.168 |
|||
| 0.164 |
0.1612 |
2303 |
|
20080917
2D Gel - Gigas 24hr Vibrio Exposure, Vibrio 24hr gigas exposure
Samples and Prep for IPG strip (ZOOM IPG pH 3-10 NL) rehydration O/N:
1. V.tubi + autoclaved oysters 24hr from 20080828 ; extracted by Tatyana 20080915 - 20uL + 135uL sample rehydration buffer.
2. V. tubi in sterile SW 24hr from 20080828 ; extracted by Tatyana 20080915 - 40uL + 115uL sample rehydration buffer.
3. Gigas gill 24hr control from 20080828 , extracted today - 4.68uL (15ug) + 150.32uL sample rehydration buffer.
4. Gigas gill 24hr V. tubi exposure from 20080828 , extracted today - 4.85uL (15ug) + 150.15uL sample rehydration buffer.
5. Gigas gill 24hr control extract from DATE - pool of 10 oyster samples, 1uL each sample = 15ug protein; 10uL total + 145uL sample rehydration buffer.
6. Gigas gill 24hr Vibrio exposure extract from DATE - pool of 10 oyster samples, 1uL each sample = 15ug protein; 10uL total + 145uL sample rehydration buffer.
Protein Extraction - Gigas gill control/V. tubi exposure 24hr from 20080828
Gigas gill 24hr control - 0.25g homogenized in 500uL Cell Lytic, brought volume up to 1mL after homogenization
Gigas gill 24hr V. tubi exposure - 0.23g homogenized in 500uL Cell Lytic, brought volume up to 1mL after homogenization
Samples were spun max speed 4C 15 mins. Supe transferred to new tube. Supe spun 4C 15 mins. Supe transferred to fresh tube and then quantitated using the Pierce Coommassie Bradford assay following the microplate procedure.
Concentrations were detemined using the following equation:
[sample] = (Avg. OD595 + 0.0403)/0.00007
Equation was previously determined from a set of BSA standards.
| SAMPLE |
OD595 nm |
Avg. OD595 |
[sample] (ug/mL) |
| Control |
0.186 |
||
| 0.181 |
|||
| 0.185 |
0.184 |
3204 |
|
| Exposed |
0.177 |
||
| 0.178 |
|||
| 0.174 |
0.176 |
3090 |
|
SDS/PAGE - Vibrio samples from 20080828
Samples and Prep (samples 1-6 extracted by Tatyana 20080915) :
1. Diluted Vt Autoclaved oyster 24hr - 20uL extract + 20uL 2x Sample Reducing Buffer
2. Diluted Vt H2O 24hr - 20uL extract + 20uL 2x Sample Reducing Buffer
3. Diluted Vt Live oysters 24hr - 20uL + 20uL 2x Sample Reducing Buffer
4. Vt Autoclaved oyster 24hr (extracted in pBAD lysis buffer, 500uL) - 20uL extract + 20uL 2x Sample Reducing Buffer
5. Vt H2O 24hr (extracted in pBAD lysis buffer, 500uL) - 20uL extract + 20uL 2x Sample Reducing Buffer
6. Vt Live oysters 24hr (extracted in pBAD lysis buffer, 500uL) - 20uL extract + 20uL 2x Sample Reducing Buffer
7. Vt MB pH=8.6 - 200uL 1x Sample Reducing Buffer (prepped by Sam 20080917)
8. Vt SW pH=8.6 (Resuspended in 100uL Mammalian Lysis Solution + Protease Inhibitor Cocktail, boiled 5 mins., spun max speed 4C 5 mins. Supe transferred to new tube) - 20uL extract + 20uL 2x Sample Reducing Buffer (prepped by Sam 20080917)
Samples were boiled for 5 mins. and then centrifuged max speed 4C for 5 mins. 30uL of each samples were loaded on Pierce 4-20% Tris-Hepes gel and run @ 150V ~45 mins. Stained with Coommassie for 45 mins. Destained with 10% acetic acid. Gel is loaded in the order listed above, from left to right.
 |
| external image 2867864081_06fc7f1998.jpg?v=0 |
Results: Bands are visible.
20080916
2D Gels - Gigas 24hr Vibrio Exposure
Performed IEF of samples from 20080915 . Modified voltage step by running final step @ 2000V for 45 mins. (instead of 30 mins.) in hopes of improving spot resolution. Dye fronts migrated at different rates, despite the fact that the two samples should be identical. Washed strips in 10 mL of reducing and alkylation buffers 15 mins. each with shaking. Transferred strips (with positive end closest to ladder) to Invitrogen 4-20% Tris-Glycine ZOOM gels, overlaid strips with ~400 uL of 0.5% agarose (in tris-glycine buffer) and allowed to solidify for 15 mins. Loaded 10uL of SeeBlue protein ladder. Ran gels @ 125V for 90 mins., but dye migration was only half way down the gel. Possibly due to running two gel boxes (Tatyana was running samples today, too) at one time (i.e. 125V split between the two boxes instead of 125V for each box)? Extended run time an additional 60 mins. until dye migrated to bottom of gels. Air bubbles had accumulated between the gel and plastic molds during this time. Doesn't appear to be an noticeable effect, but the gels were "stickier" and slightly difficult to remove from plastic. One gel tore in multiple places, but remained intact. (Side note: Knotched corner above ladder on Tatyana's gel that was unmarked. This gel also had the negative end of the IEF strip next to the ladder.)Stained according to Invitrogen Silverquest protocol.
One gel ripped during incubations.
Gel #1
| external image 20080916-01.JPG |
Gel #2
| external image 20080916-02.JPG |
Results: The two gels above are duplicates. They should appear to be exactly the same, but do not. Getting both horizontal and vertical streaking.
Bacterial Cultures
Inoculated 5mL LB+Kan with colonies from pCR2.1/COX (Colony #6), pBAD/Decorin, FST-L, LAP cloning for miniprep tomorrow. Incubated O/N, 37C, 250RPM.
20080915
2D Gels - Gigas 24hr Vibrio Exposure
Loaded duplicate samples (15ug) of C. gigas 24hr Vibrio exposure gill protein extracts (VE1-10 pooled from 20080617) onto two IEF strips (Invitrogen ZOOM IPG pH 3-10NL) for rehydration O/N.20080912
 |
| Picture_3.png |
20080911
Real-time PCR - V.tubiashii pH exp. and tank exp.
Performed real-time PCR with V.tub 16s primers (original set) using 2x Immomix on Tatyana's cDNA, diluted 1:4. This is a repeat of what she had done due to the presence of a signal in her water sample on 20080910. This was run over night. PCR set up/plate layout here .Samples:
pH-6.62
pH-7.57
pH-8.57
pH-9.56
Vt + auto gigas t=24h
Vt + live gigas t=24h
gigas only t=24h
Vt + sea water t=24
Results: Got an acceptable 16s signal in all samples except the oyster only sample from the tank experiment. No signal in water samples.
PCR - All Sea Fan samples and Chelex Supernatant Samples
50uL of each sea fan sample (now referred to as SUPE) was transferred to a new tube. 250uL of 10% Chelex was added to each of these and vortexed. All samples (original and SUPE) were boiled for 30 mins. and then spun at 4C for 30 mins. PCR set up is here (titled "PCR LABY Samples All"). PCR reaction ran overnight. NOTE: "Old" references the four samples provided by Steven that only have a number on them and no other information. "New" refers to the newer samples provided by Steven that are labeled as either "Spot Fan" or "Sea Fan" and a number.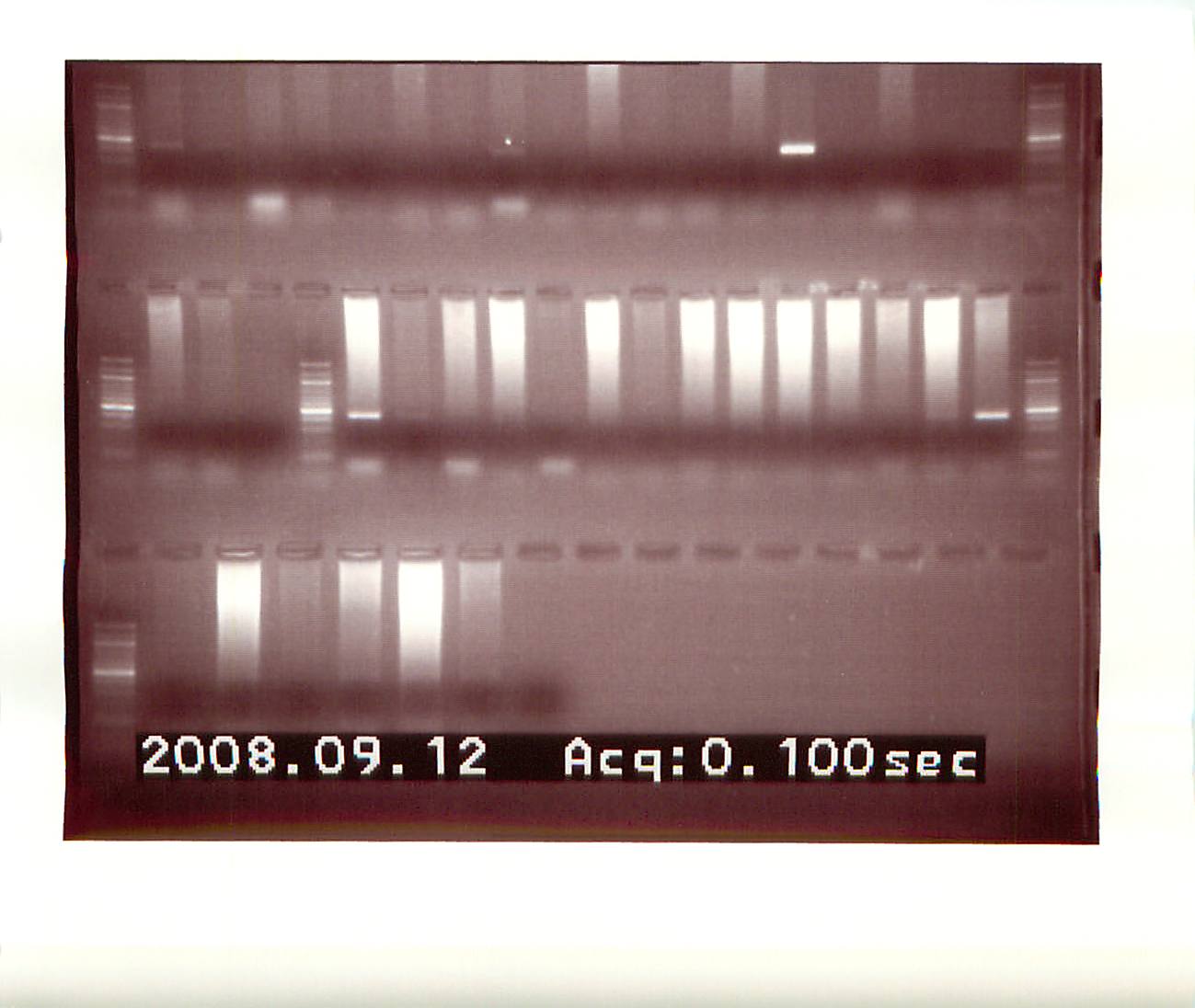 |
| external image 20080912.jpg |
TOP
Lane 1 = 100bp Ladder
Lane 2 = Old #1
Lane 3 = Old #4
Lane 4 = Old #6
Lane 5 = Old #8
Lane 6 = Control Fan 1
Lanes 7-19 = Sea/Spot fan samples 1-13
Lane 20 = 100bp Ladder
MIDDLE
Lane 1 = 100bp Ladder
Lanes 2-3 = Sea/Spot fan samples 14-15
Lane 4 = Neg. Control
Lane 5 = 100bp Ladder
Lane 6 = Old #1 SUPE
Lane 7 = Old #4 SUPE
Lane 8 = Old #6 SUPE
Lane 9 = Old #8 SUPE
Lane 10 = Control Fan 1 SUPE
Lanes 11-19 = Sea/Spot fan samples 1-9
Lane 20 = 100bp Ladder
BOTTOM
Lane 1 = 100bp Ladder
Lanes 2-7 = Sea Spot fan samples 10-15
Lane 8 = Neg. Control
Results: Bands were cut and purified with Millipore spin columns from:
Old #1, Old #4, New #3, New #9, New #13, Old #1 SUPE, Old #4 SUPE, New #8 SUPE, New #9 SUPE, New #13 SUPE, New #14 SUPE, New #15 SUPE
PCR - Sea Fan samples w/LABY Primers w/varying [Mg2+]
Samples were vortexed. Boiled samples 30mins. Spun 30mins. @ 4C. PCR set up here.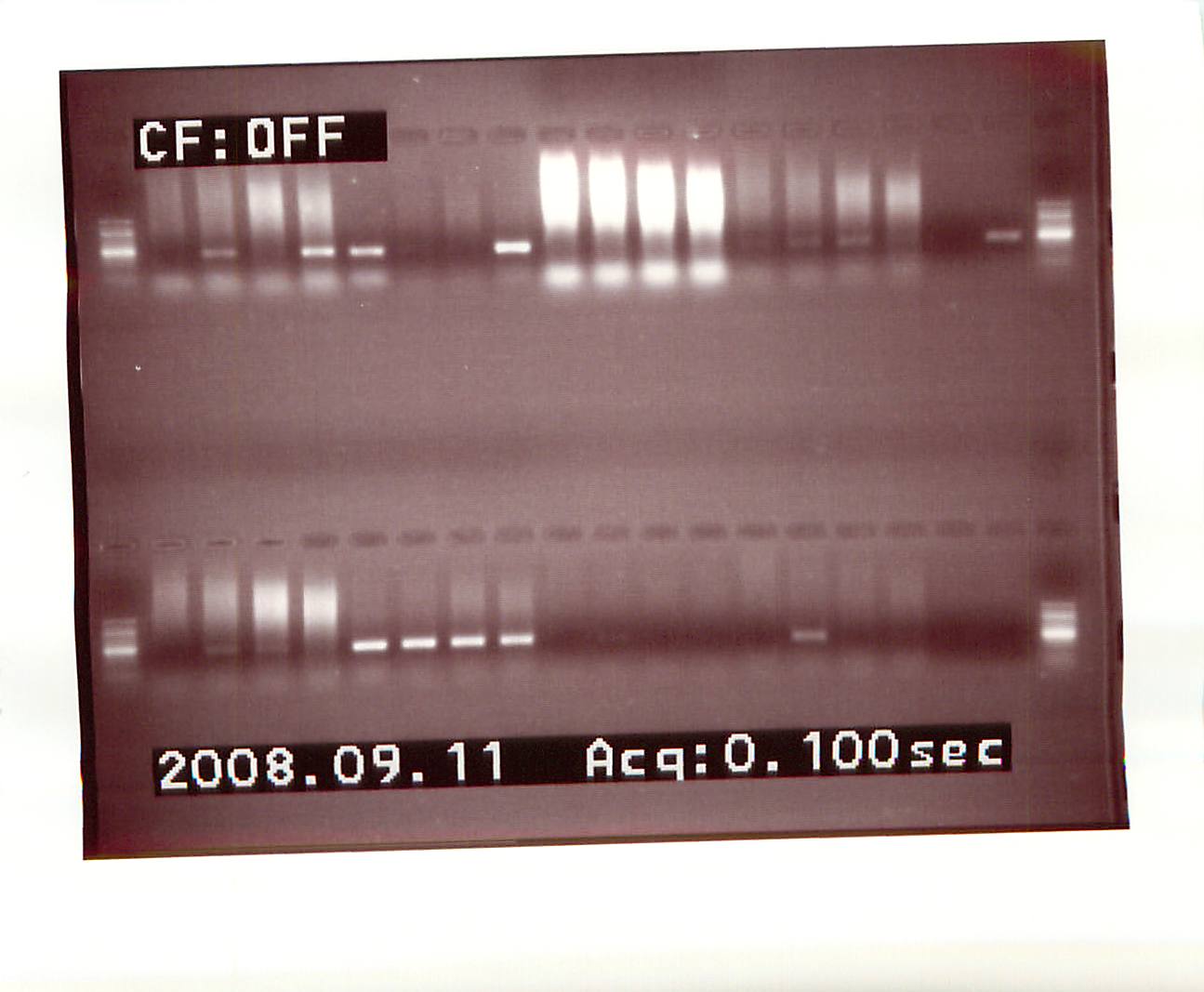 |
| external image 20080911.jpg |
Lane 1 = 100bp Ladder
Lanes 2-5 = Old #1 - 1.5mM, 2mM, 3mM, 4mM
Lanes 6-9 = Old #4 - 1.5mM, 2mM, 3mM, 4mM
Lanes 10-13 = Old #6 - 1.5mM, 2mM, 3mM, 4mM
Lanes 14-17 = Old #8 - 1.5mM, 2mM, 3mM, 4mM
Lane 18 - 1.5mM Neg. Control
Lane 19 - 2.0mM Neg. Control
Lane 20 - 100bp Ladder
BOTTOM HALF:
Lane 1 = 100bp Ladder
Lanes 2-5 = New #1 - 1.5mM, 2mM, 3mM, 4mM
Lanes 6-9 = New #9 - 1.5mM, 2mM, 3mM, 4mM
Lanes 10-13 = New #10 - 1.5mM, 2mM, 3mM, 4mM
Lanes 14-17 = Contr. #1 - 1.5mM, 2mM, 3mM, 4mM
Lane 18 - 3.0mM Neg. Control
lane 19 - 4.0mM Neg. Control
Lane 20 - 100bp Ladder
Results: Apparent contamination in 2.0mM preps, as a band is present in Top Half - Lane #19 (2.0mM Neg. Control). Bands were cut from Bottom Half - Lanes 4 & 8 (New 1 and New 9, 3mM, respectively) and purified using Millipore spin columns.
20080903
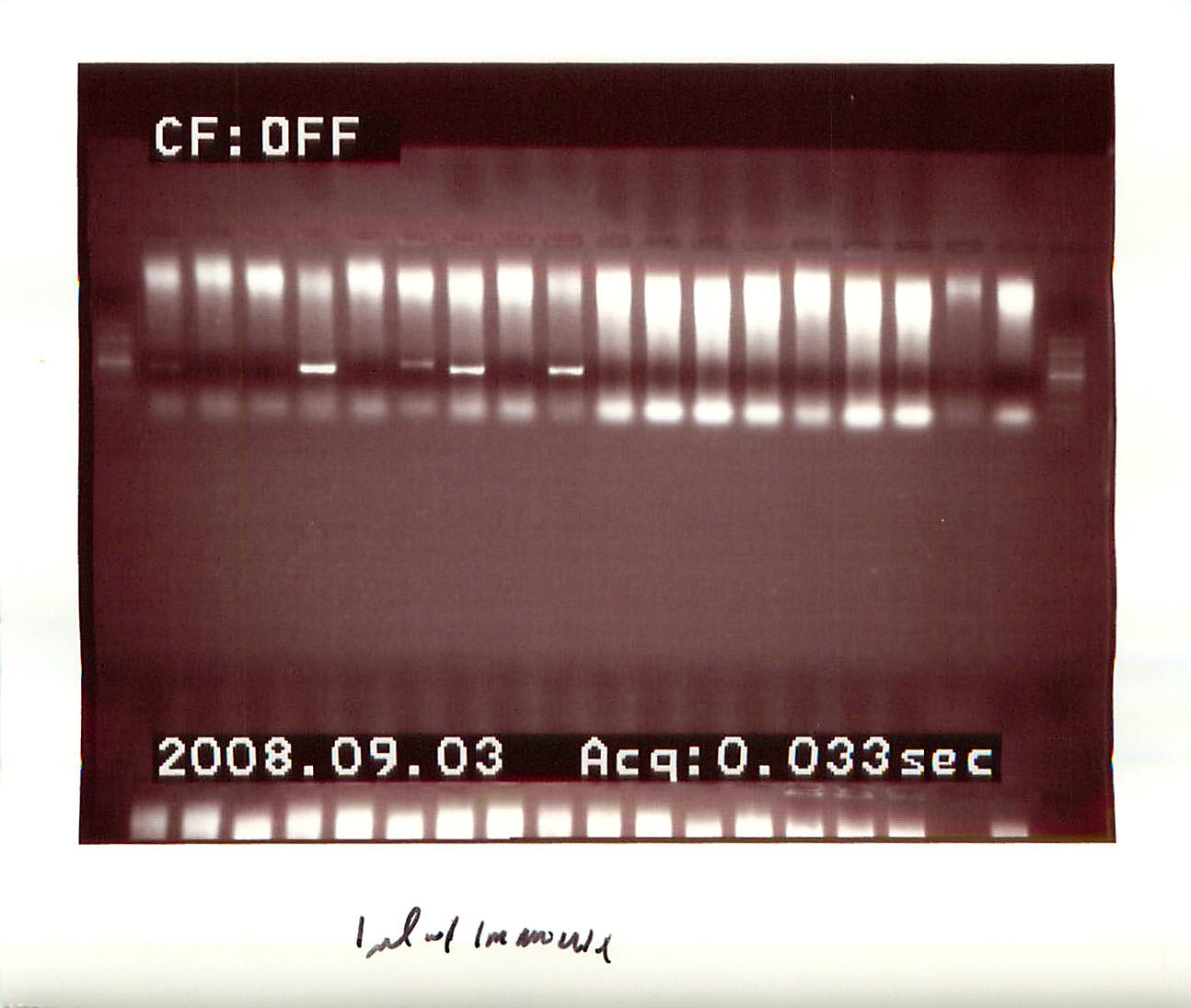 |
| external image 20080903-01.jpg |
 |
| external image 20080903-02.jpg |
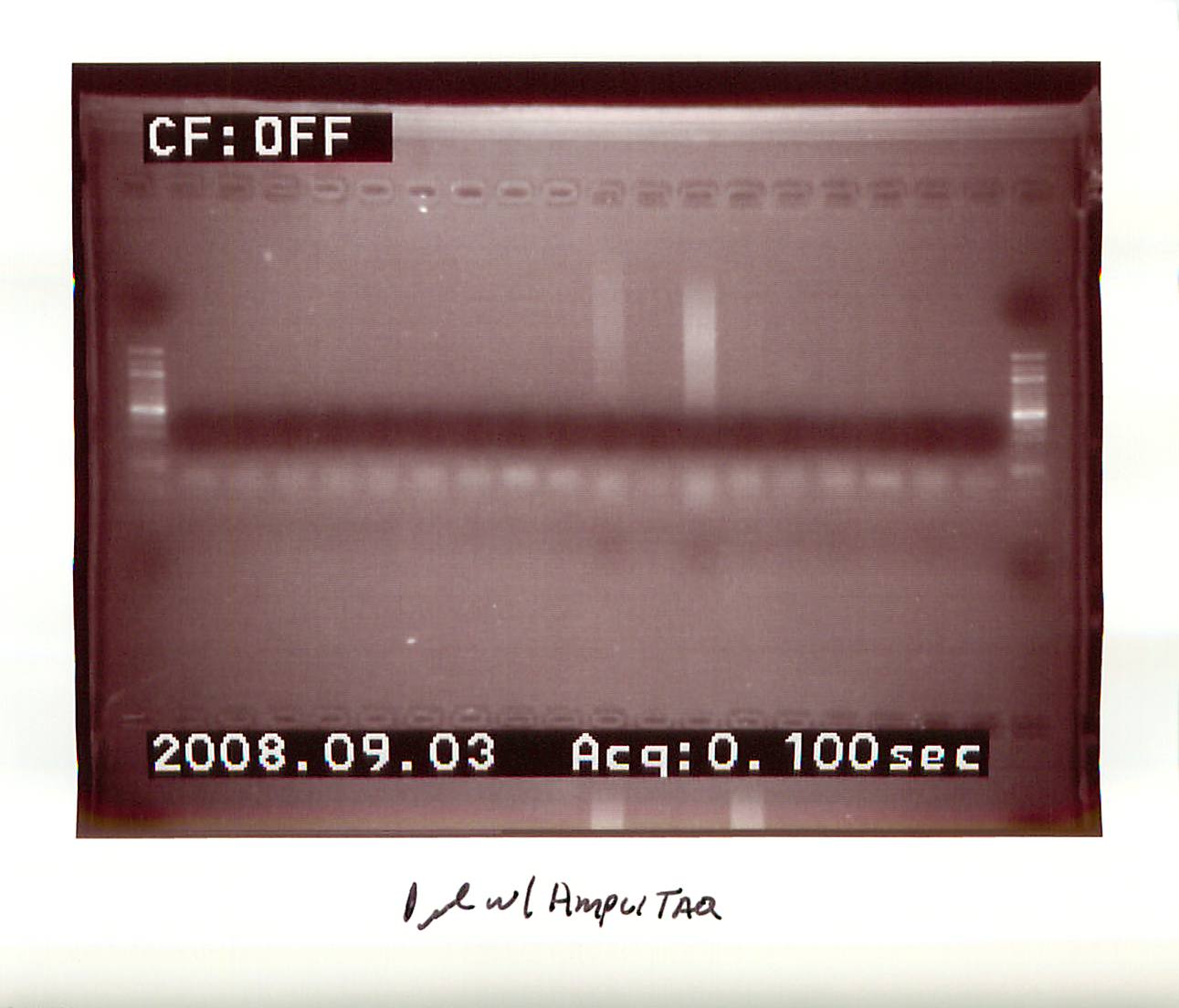 |
| external image 20080903-03.jpg |
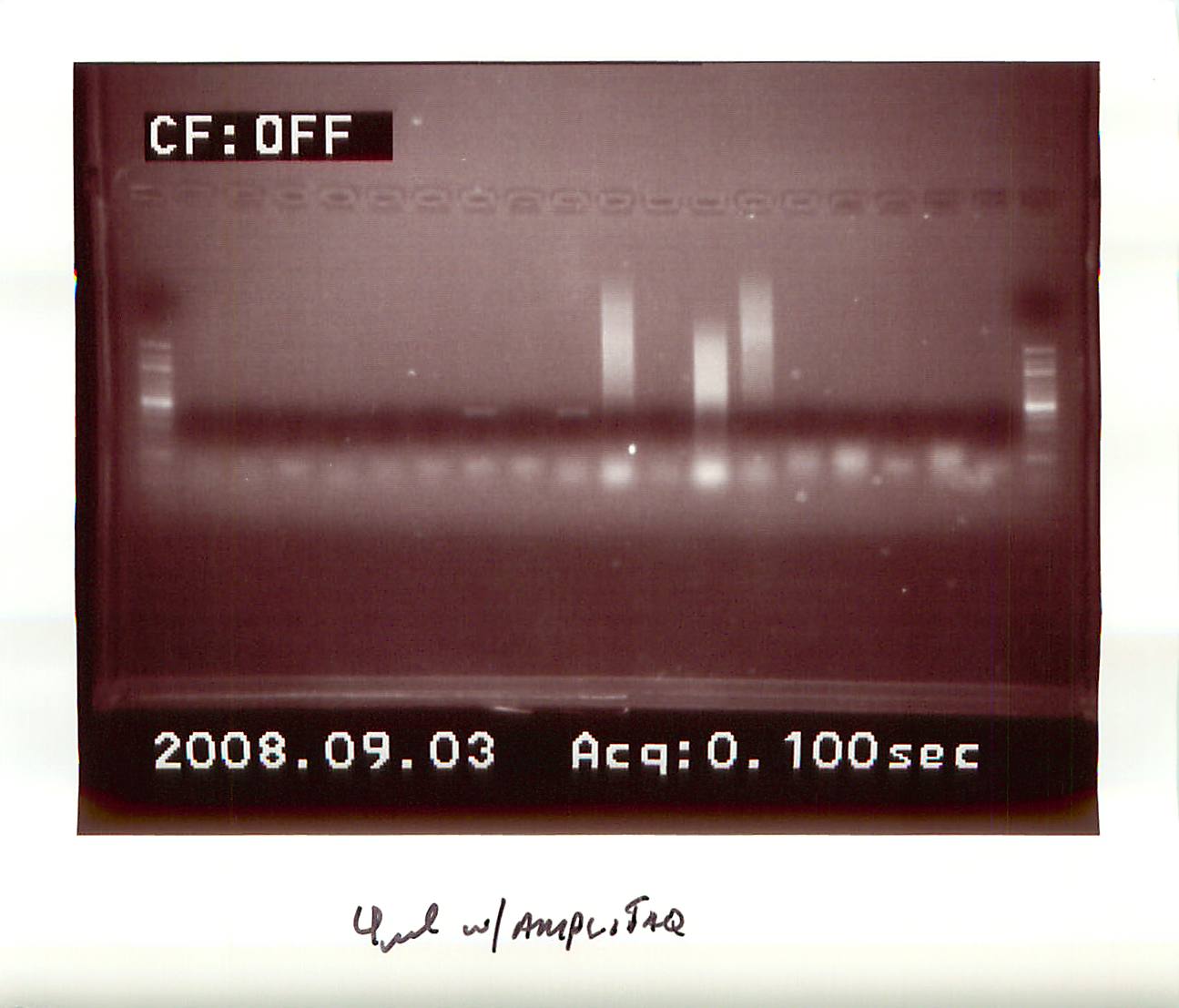 |
| external image 20080903-04.jpg |
20080902
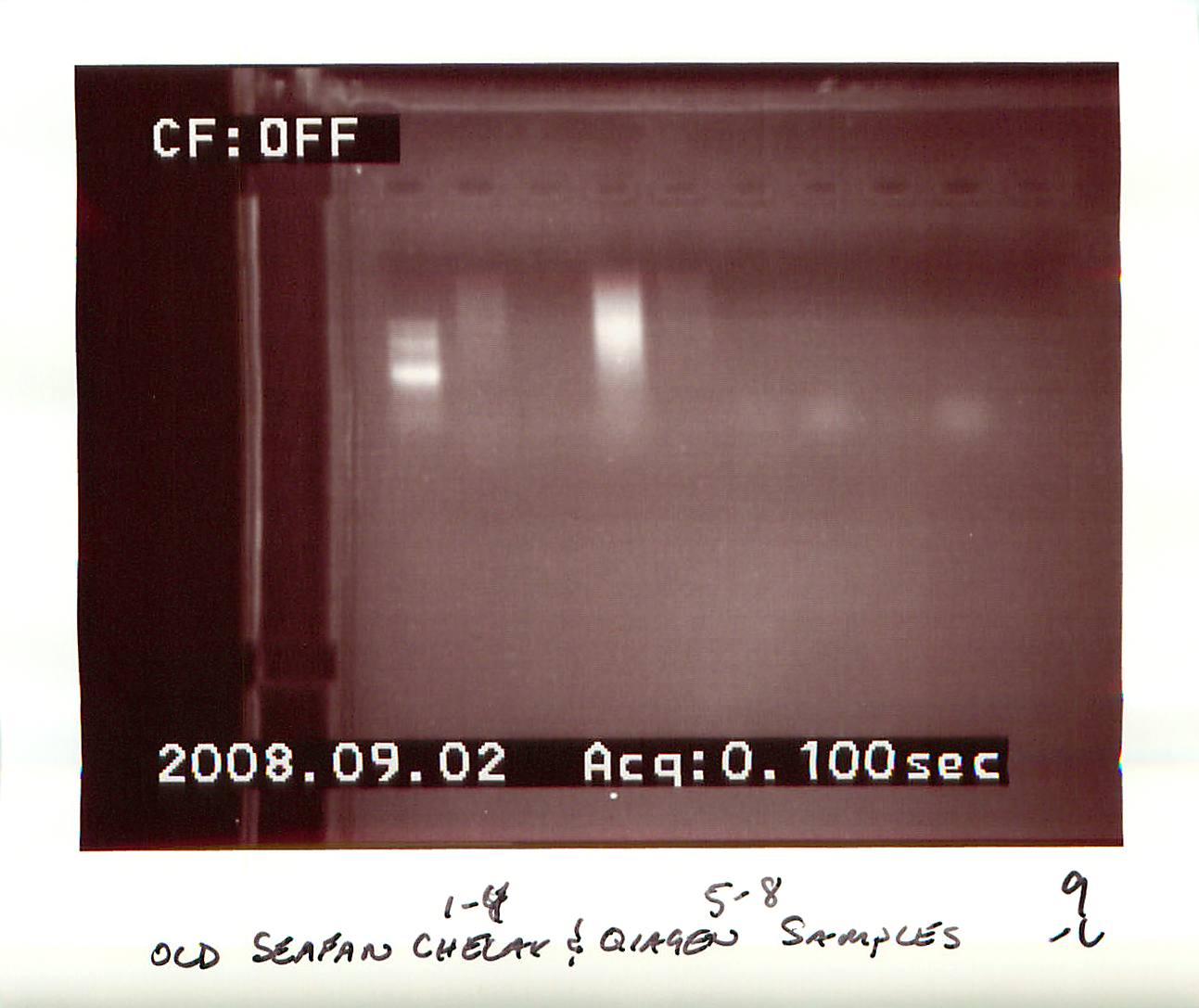 |
| external image 20080902.jpg |
20080828
 |
| external image 20080828.jpg |
20080820
 |
| external image 20080820-01.jpg |
 |
| external image 20080820-02.jpg |
20080814
 |
| external image 20080814-01.jpg |
 |
| external image 20080814-02.jpg |
20080812
Insert gel #1 here.
 |
| external image 20080812-02.jpg |
20080811
 |
| external image 20080811-01.jpg |
 |
| external image 20080811-02.jpg |
 |
| external image 20080811-03.jpg |
20080806
PCR - Products for pBAD cloning
PCRs were performed on previously cloned inserts in pGAD, unless noted.
Primers used:
Decorin:
FST:
FST-L:
LAP:
Telethonin:
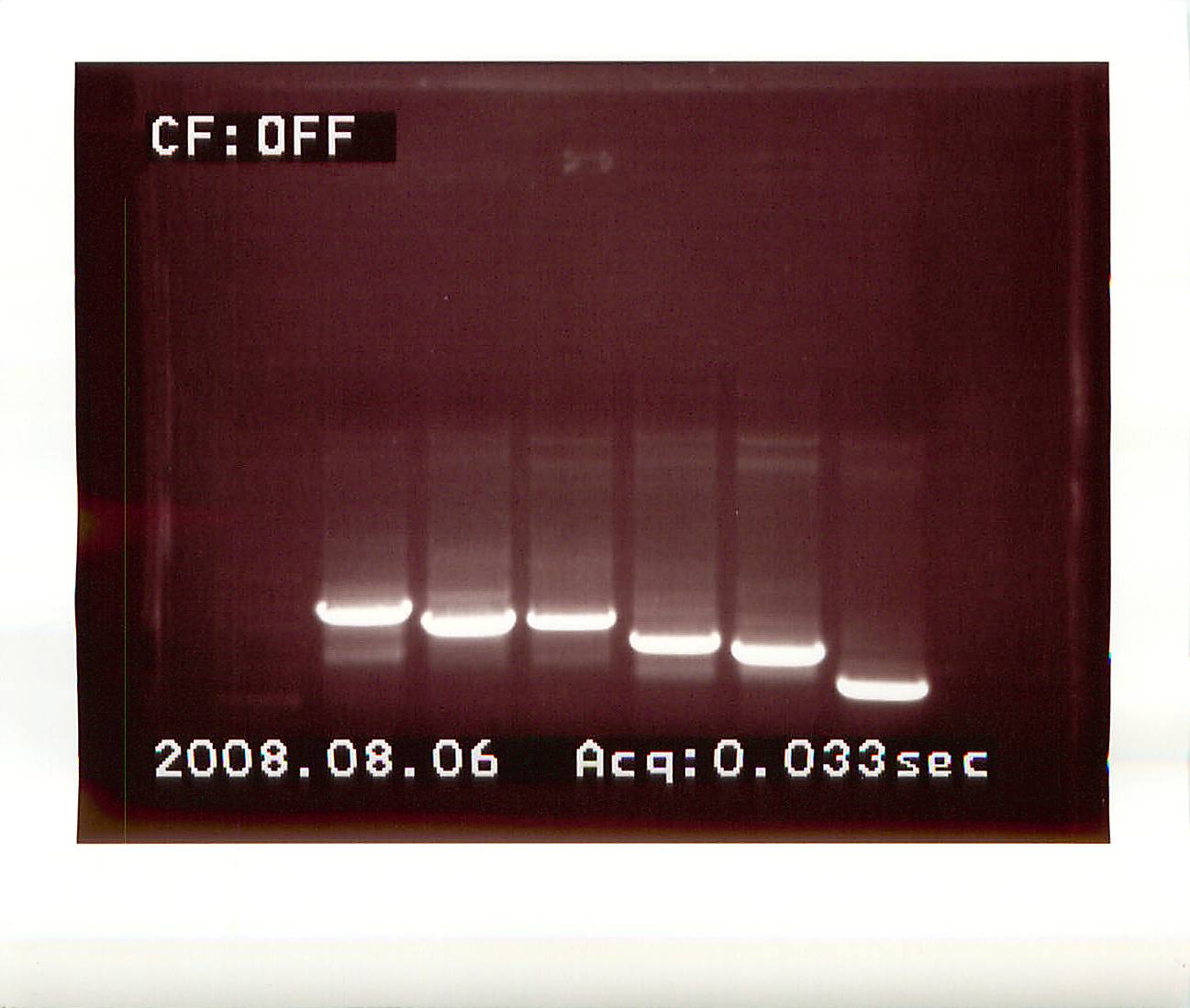 |
| external image 20080806%20.jpg |
| Lane |
Sample |
Notes |
| 1 |
100bp ladder |
|
| 2 |
Decorin |
|
| 3 |
FST |
|
| 4 |
FST (from pGBK) |
|
| 5 |
FST-L |
|
| 6 |
LAP |
|
| 7 |
Telethonin |